- Title
-
Zebrafish as a Model for the Study of Live in vivo Processive Transport in Neurons
- Authors
- Bercier, V., Rosello, M., Del Bene, F., Revenu, C.
- Source
- Full text @ Front Cell Dev Biol
|
Construct expression and cell type selection. |
|
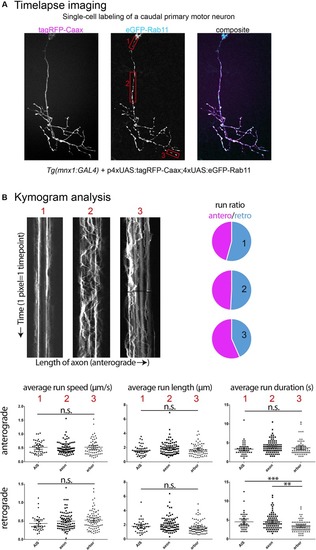
Examples of time-lapse analysis. |