- Title
-
Comparative transcriptome analysis reveals the role of p53 signalling pathway during red-spotted grouper nervous necrosis virus infection in Lateolabrax japonicus brain cells
- Authors
- Xiang, Y., Jia, P., Liu, W., Yi, M., Jia, K.
- Source
- Full text @ J. Fish Dis.
|
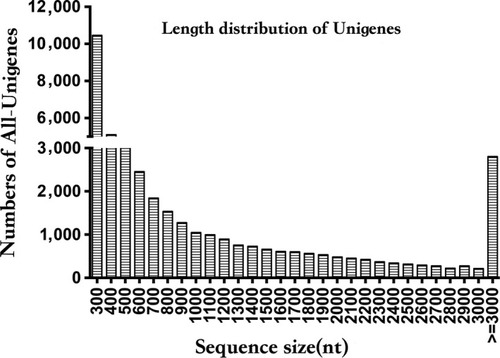
Length distribution of All‐unigenes |
|
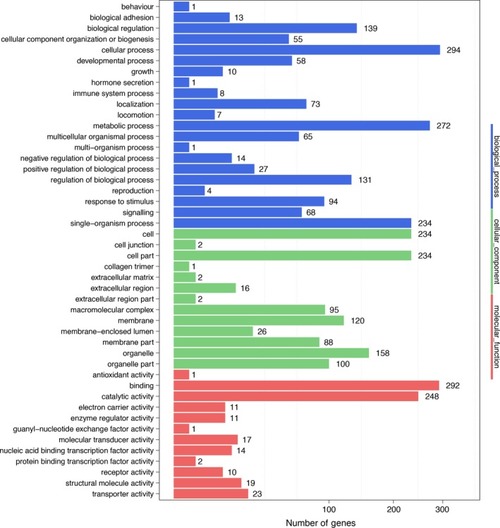
Gene Ontology function classification of All‐unigenes [Colour figure can be viewed at |
|
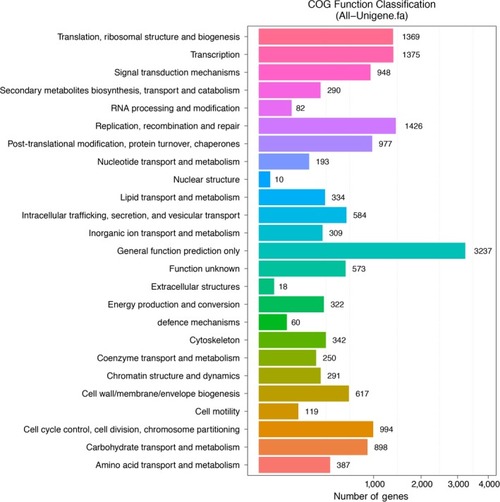
Clusters of Orthologous Groups (COG) function classification of All‐unigenes [Colour figure can be viewed at |
|
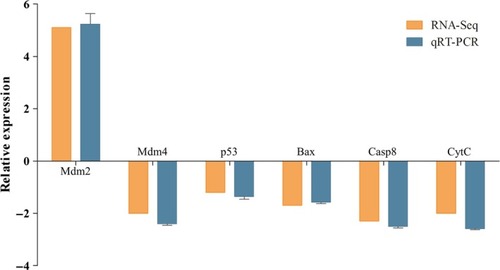
Comparison of the fold change expression of six differentially expressed genes involved in p53 signalling pathway as determined by RNA‐seq and qPCR. The results of qPCR were evaluated by normalizing to |
|
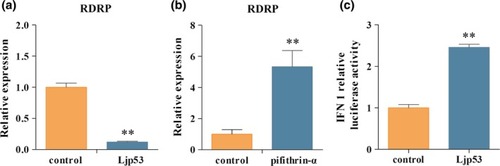
Effects of Ljp53 on RGNNV replication and zebrafish IFN1 promoter activity. (a) qRT‐PCR detection of |
|
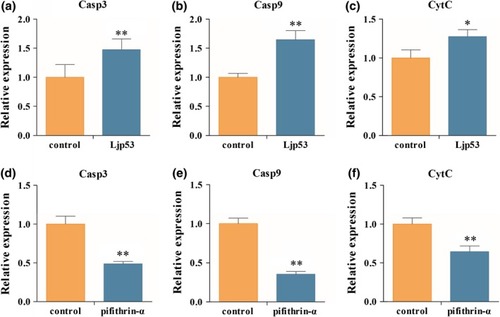
Expression of apoptosis‐related genes. qRT‐PCR detection of |