- Title
-
Distinct regulatory mechanisms act to establish and maintain Pax3 expression in the developing neural tube
- Authors
- Moore, S., Ribes, V., Terriente, J., Wilkinson, D., Relaix, F., Briscoe, J.
- Source
- Full text @ PLoS Genet.
|
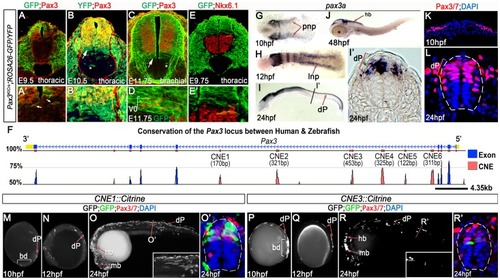
Pax3 expression is recapitulated by the activity of 2 CRMs. (A, A′) At E9.5, Pax3 expression correlates with cells labeled by YFP in Pax3Cre/+;Rosa26-YFP mouse embryos (n = 2). However between E10.5 (B) (n = 2) and E11.75 (C) (n = 2), cells derived from the Pax3 lineage are found in the both the dorsal and ventral neural tube of Pax3Cre/+;Rosa26-GFP/YFP mouse embryos. In the ventral neural tube, clusters of isolated cells (asterisk in B and C) and a domain spanning 3–4 cells adjacent to the Pax3 ventral boundary (B′ and arrows in B and C) is observed. (D) This domain of transgene expression encompasses, but is not limited to, Evx1 expressing ventral interneurons at E11.75 (n = 2). (E, E′). Consistent with the labeling of interneurons at E11.75, embryos assessed at E9.75 exhibit a boundary of Pax3 lineage opposed to the dorsal limit of the Nkx6.1 domain (n = 3). (F) Comparative genomic analysis of the Pax3 locus reveals 6 CNEs within the 4th intron of the gene. (G) Pax3a mRNA is first detected in the developing midbrain, hindbrain and intermediate regions of the posterior neural plate (pnp) of zebrafish embryos at 10 hpf. (H) By 12 hpf, transcription is limited to the lateral neural plate (lnp) posteriorly. (I) At 24 hpf, pax3a is highly expressed in the midbrain, hindbrain and progenitors within the dorsal spinal cord (dP), as shown in (I′). (J) Transcription is rapidly downregulated in the spinal cord after 24 hpf and is not detectable at 48 hpf. (K–L) Pax3/7 protein, visualised with the DP312 antibody, is also restricted to the intermediate and lateral regions of the neural plate at 10 hpf and the dorsal half of the spinal cord at 24 hpf. (M–N) Profile views of CNE1::Citrine transient transgenic embryos at 10 and 12 hpf showing enhancer activity in the tail bud (bd) and lateral neural plate (M) (n = 25/27), then within dorsal neural progenitors (N) (n = 19/22). (O, O′) At 24 hpf, CNE1::Citrine transient transgenics recapitulate pax3a expression across the AP axis of the CNS and the DV axis of the spinal cord. (P–R′) CNE3 is active in the neural plate and tail bud at 10 and 12 hpf (n = 10/10, n = 7/7), however embryos rarely exhibit labeling of Pax3/7 expressing progenitors within the dorsal spinal cord at 24 hpf (n = 5/48). |
|
CNE3 balances transcriptional activation and repression to establish the Pax3 expression domain. (A) TCFsiam transgenic zebrafish assessed at 10 hpf reveal activated Wnt signaling in the posterior neural plate and tailbud (bd). (A′) Transverse sections of the posterior neural plate demonstrate that Wnt pathway activity is not restricted in the medio-lateral axis of the tissue, whereas Pax3/7 is expressed laterally. (B–C) Wnt signaling is maintained within the neural tube up to 18 hpf, following which it rapidly declines and is found only in the dorsal most row of cells within the neural tube at 24 hpf. (D) Matrix indicating the conservation of CNE3-Motif5 across 12 vertebrate genomes. (E, E′) Zebrafish injected with a CNE3 reporter rarely contain labeled cells within the dorsal spinal cord of transient transgenic embryos at 24 hpf (n = 4/33) and the reporter is not active in the chick neural tube at E3 (F) (n = 0/7). (G, G′) CNE3Motif5Del::Citrine transgenics exhibit an increase of enhancer activity across the DV axis of the spinal cord, compared to controls (n = 28/32, p<0.0001). (H) Furthermore, deletion of Motif5 results in the ectopic induction of CNE3 activity in chick embryos at E3 (n = 4/4, p = 0.003). (I) Schematic depicting the organisation of motifs within CNE3 and the mutation induced within the conserved HD binding site in Motif5. (J, J′) Mutation of the HD binding site in Motif5 increases CNE3 activity in progenitors within the zebrafish spinal cord, compared to controls (n = 21/21, p<0.001). (K) Chick embryos electroporated with CNE3M5HDMut DNA exhibit ectopic enhancer activity in the developing neural tube (n = 3/4, p = 0.0242). (L) EMSAs performed using a DNA probe spanning Motifs 3–5 of CNE3 and chick spinal cord nuclear extract. Complexes formed with nuclear extract are indicated (arrows, compare Control to Probe lanes). Addition of an Nkx6.1 antibody creates a slower migrating DNA/protein complex than controls, whereas addition of FoxA2 antibody abrogates complex formation. Both results indicate that these proteins are able to bind CNE3. Addition of Dbx1, Dbx2, Pax6 and Nkx6.2 antibodies have a minor effect on the motility of EMSA complexes. (M, M′) Consistent with binding and mutagenesis studies, the electroporation of Nkx6.1 in the chick neural tube is sufficient to repress endogenous Pax3 protein expression in chick embryos (n = 11). |
|
Functional dissection of CNE1 enhancer activity. (A) Schematic outlining the organisation of motifs within CNE1. (B, B′) CNE1 transient transgenic zebrafish embryos recapitulate pax3a expression across the AP axis of the CNS, including the spinal cord (n = 66/67). (C) Zebrafish CNE1 sequence specifically drives transgene expression in the Pax3/7 domain of the chick spinal cord, despite widespread transfection of LacZ across the DV axis of the tissue (n = 10/12). (D) Matrix representing the degree of Motif1 conservation across 12 vertebrate genomes. (E, E′) Deletion of Motif1 results in a complete loss of CNE1 activity in zebrafish spinal cord at trunk level (n = 0/22, p<0.0001), however the enhancer remains active in the anterior CNS (n = 22/22) and the most posterior region of the spinal cord (n = 8/22). (F) Loss of Motif1 greatly reduces CNE1 activity in the chick neural tube (n = 1/8, p = 0.0019). Motif2 (G), is not required for CNE1 activity in the zebrafish (H, H′) (n = 37/38) or chick (I) (n = 4/6) spinal cord. Loss of Motif 3 (J) reduces CNE1 mediated transcription across the AP axis of the zebrafish CNS (K, K′) (n = 0/26, p<0.0001) and precludes activity in the chick spinal cord (L) (n = 0/5, p = 0.003). Deletion of Motif4 (M) does not significantly alter the activity of CNE1 in zebrafish (N, N′) (n = 20) or chick (O) (n = 4/5). |
|
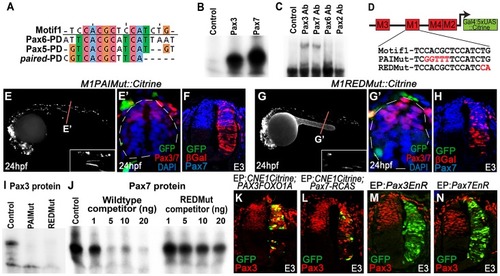
CNE1 mediates direct autoregulation and positive feedback via a paired domain binding site. (A) CNE1-Motif1 is homologous to the 5′ region of defined PD binding sites, however the alignment diverges in the 3′ region. (B) EMSA performed using Motif1 DNA and in-vitro synthesised Pax3 and Pax7 proteins, both of which can bind the sequence. (C) EMSA using Motif1 DNA and chick spinal cord nuclear extract, addition of antibodies against Pax3 or Pax7 to the reaction decreases the mobility of the DNA/protein complex. Addition of Pax6 or Pax2 antibodies does not alter the distribution of complexes within the EMSA. (D) Schematic illustrating the mutations targeted within CNE1-Motif1. (E, E′) M1PAIMut transgenic zebrafish exhibit a marked reduction in CNE1 activity in spinal cord progenitors, compared to the wildtype enhancer (n = 2/38, p<0.001). (F) Mutation of the PAI domain binding site precludes CNE1 activity in the chick neural tube (n = 0/8, p = 0.0007). (G, G′) Mutation of the RED domain binding site phenocopies Motif1 deletion in zebrafish embryos (n = 0/19, p<0.001) and chick embryos (H) (n = 1/5, p = 0.0128). (I) EMSAs performed using in vitro synthesised Pax3 protein and Motif1 DNA harboring either PAI or RED mutations. Mutation of either PAI or RED region precludes complex formation. (J) Competition EMSA performed using in vitro synthesised Pax7 protein, radiolabelled wildtype Motif1 DNA and non-labeled competitor probes. Non-labeled wildtype DNA effectively competes with radiolabelled probe for Pax7 binding, whereas DNA harboring the RED mutation cannot. (K) Electroporation of PAX3FOXO1A-RCAS induces ectopic CNE1 activity and Pax3 protein expression in the ventral neural tube (n = 9). (L) Pax7-RCAS electroporation also induces CNE1 activity and Pax3 expression (n = 7). Electroporation of dominant negative forms of Pax3 (M) (n = 7) or Pax7 (N) (n = 6) represses Pax3 protein expression within its endogenous domain. |
|
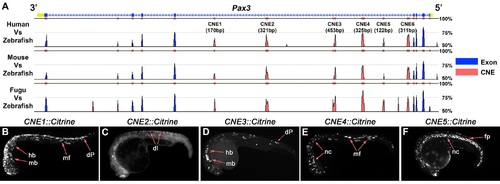
Several functional CRMs are located within the 4th intron of the Pax3 locus. (A) The full Mulan alignment of the Pax3 locus, summarised in Figure 1E. (B) CNE1 transient transgenics exhibit reporter expression across the AP axis of the developing CNS at 24 hpf (n = 67). (C) By contrast, CNE2 activity weakly labels postmitotic neurons within the dorsal spinal cord (n = 11). (D) At 24 hpf, CNE3 is active within the midbrain (mb), hindbrain (mb) and progenitors within the dorsal spinal cord (dP) (n = 51). (E) CNE4 is sufficient to direct transcription within muscle fibres (mf) and cranial neural crest (nc) (n = 32). (F) Surprisingly, CNE5 robustly labels the notochord (nc) and floor plate (fp) of the neural tube, tissues that do not express Pax3 at any point of their development (n = 34). |
|
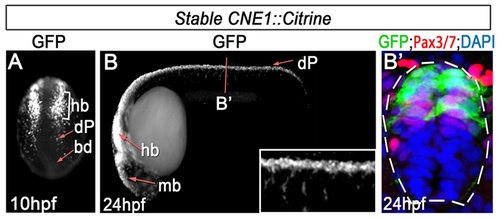
CNE1 activity recapitulates Pax3 expression. (A) CNE1 stable transgenic embryos assessed at 10 hpf exhibit Citrine expression in the developing hindbrain (hb) and presumptive dorsal progenitors (dP) within the lateral regions of the posterior neural plate. (B, B′) At 24 hpf, CNE1 activity recapitulates pax3a expression across the AP axis of the CNS and is restricted to the Pax3/7 domain of the dorsal spinal cord. |
|
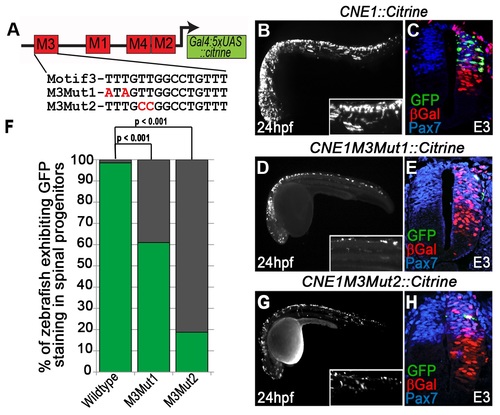
Mutation of the HMG box site within Motif3 reduces CNE1 activity. (A) Schematic outlining the mutations introduced into the HMG box site of Motif3. M3Mut1 transgenic zebrafish exhibit a reduction in CNE1 activity compared to controls (compare B to D) (n = 25/41, p<0.001). A similar reduction in CNE1 activity is observed in M3Mut2 transgenics (compare B to G) (n = 6/32, p<0.001). (F) Graphical summary of the effect of HMG binding site mutations upon CNE1 activity in zebrafish embryos. Experiments performed in chick reveal a reduction in CNE1 activity in both M3Mut1 (n = 3/7) and M3Mut2 (n = 2/5) electroporations, however this result did not reach statistical significance (compare C to E and H, respectively). |