- Title
-
Initiation of cyp26a1 Expression in the Zebrafish Anterior Neural Plate by a Novel Cis-Acting Element
- Authors
- Chen, C., Stedman, A., Havis, E., Anselme, I., Onichtchouk, D., Giudicelli, F., Schneider-Maunoury, S.
- Source
- Full text @ PLoS One
|
Characterization of cANE activity as an early neural plate specific enhancer.(A-I,A′-I′) Compared expression patterns of cyp26a1 (A-I) and egfp driven by cANE (A′-I′) during early embryonic development. (J) Double in situ hybridization showing barhl2 expression domain (blue) exactly filling the gap in the cyp26a1 expression domain (red). (A,B,C,E,G,A′,B′,C′,E′,G′) are lateral views. (d,f,h,I,D′,f′,h′,I′) are animal pole views. White arrowheads: anterior neural plate. Black arrowheads: blastoderm marginal zone. Arrows in (H-J): gap in the anterior neural plate domain of cyp26a1 expression. All stages are indicated in the pictures. (k,k′,l,l′) The effect of 100 nM retinoic acid (RA) treatment between 2,5 hpf and 8,5 hpf on on stable transgenic cANE_endo:::egfp (K-K′) and cANE::egfp (L-L′) expression. (M) EGFP fluorescence in a 12 hpf stable transgenic cANE:::egfp embryo. Lateral view with dorsal to the left. (N) Schematic representation of cANE and all three reported retinoic acid responsive elements (R1, R2, R3) identified previously. cANE is located from -504 bp to -195 bp relative to cyp26a1 ATG codon. An: animal, Vg: vegetal; V: ventral; D: dorsal, A: anterior, P: posterior, L: left, R: right, CTRL: control embryo, RA: retinoic-acid treated embryo. |
|
Detailed dissection of cANE by transient expression of deletion constructs in zebrafish embryos.(A) Schematic view of the constructs used for dissection of the cANE module. All constructs contain one single fragment (black) from the cANE module, placed immediately upstream of the gata2 minimal promoter, except for construct cANE_endo, where the gata2 promoter is replaced by the cyp26a1 endogenous promoter. Numbers correspond to nucleotide coordinates within the 310 nt zebrafish cANE. The hatched block (Motif1) represents the 12-bp difference between constructs 39-310 and 50-310; the highly conserved blocks are indicated by checkered patterns. (B-K) Enhancer activity of the cANE deletions shown in (A), assayed by egfp in situ hybridization in transient or stable (labelled Tg()) transgenic embryos. All embryos are between stages 10.5 and 11 hpf, viewed from the animal pole, with anterior on the left. |
|
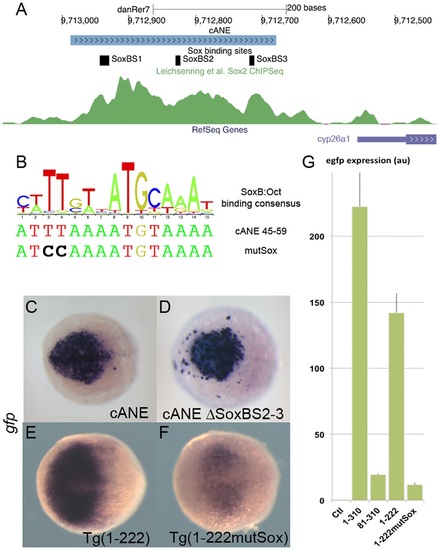
Role of SoxB binding sites in the regulation of cyp26a1 expression in the anterior neural plate (ANP).(A) Representation of the cANE region in the zebrafish genome; 3 predicted SoxB binding sites are indicated; the green channel represents the intensity of the anti-Sox2 ChipSeq signal in this region according to [34]. (B) A logo representing the composite consensus binding site for SoxB/Oct factors [33] is aligned with the predicted Sox binding site (Sox_BS1) overlapping Motif1 (cANE 45-59). The mutSox TT->CC mutation destroying the Sox-binding half-site is indicated. (C-F) egfp in situ hybridization representing enhancer activity of cANE ΔSoxBS2-3, where both Sox_BS2 and Sox_BS3 have been deleted (C), compared with intact cANE (D), at the 90% epiboly stage, and enhancer activity of Motif1 mutation mutSox (F) in 1-222 context compared with wild type 1-222 (E), at the 75% epiboly stage. Dorsal views; anterior is to the left. (G) RT-PCR relative quantification of total egfp expression stable transgenic embryos for constructs cANE (1-310), 81-310, 1-222 and 1-222 mutSox, as well as non transgenic embryos (Ctl); error bars represent SEM; units are arbitrary. |