- Title
-
Control of brain patterning by Engrailed paracrine transfer: a new function of the Pbx interaction domain
- Authors
- Rampon, C., Gauron, C., Lin, T., Meda, F., Dupont, E., Cosson, A., Ipendey, E., Frerot, A., Aujard, I., Le Saux, T., Bensimon, D., Jullien, L., Volovitch, M., Vriz, S., Joliot, A.
- Source
- Full text @ Development
|
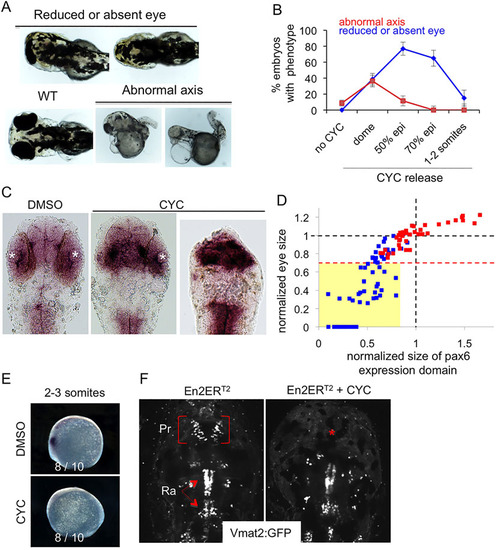
Engrailed gain of function results in reduction in both pax6 expression domain and eye size. (A-F) mRNA encoding En2ERT2 was injected at the one-cell stage and the protein was activated at different times of development. Eye size reduction (up to total disappearance) and axis abnormality phenotypes (A) were scored at different times of activation controlled by cyclofen release (B). WT, wild type; epi, epiboly. Diencephalons were measured on flat-mount embryos as the anterior pax6 expression domain (C) and compared to the eye phenotype (mean of the two eye sizes) for each embryo (D), demonstrating the correlation between these two parameters. White asterisks, eye position; red squares, control; blue squares, activated Engrailed. The dashed red line indicates the limit below which eyes were scored as reduced (or absent). All the embryos of this class (yellow box) have a small anterior pax6 expression domain. Additional effects of En2 activation were apparent at the 2-3 somite stage with the loss of the anterior pax6 expression domain (E) and at 4dpf with the selective loss of the pretectal vmat2:GFP neuronal cluster (red asterisk) (F). Pr, pretectal cluster; Ra, raphe. |
|
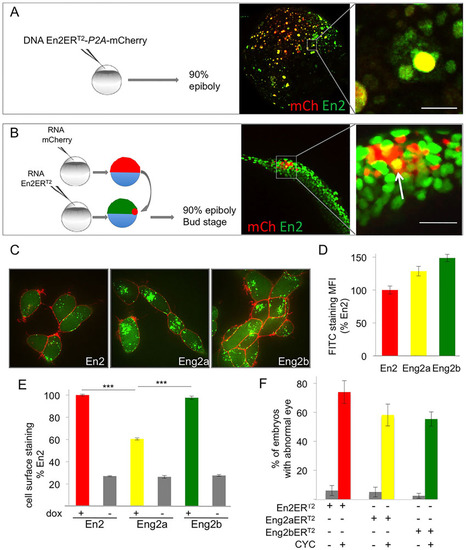
Zebrafish Engrailed 2a and 2b are able to transfer between cells in vivo and ex vivo. (A,B) In vivo intercellular transfer of En2 protein. (A) The En2ERT2-P2A-mCherry plasmid was injected at the one-cell stage (10ng/µl). Embryos were fixed at 90% epiboly and immunostained for En2ERT2 (green) and mCherry (red). En2 transfer was visualised by the presence of En2ERT2 staining (green) in non-injected cells (negative for mCherry staining). (B) Cells expressing mCherry (red) were transplanted into 30% epiboly embryos ubiquitously expressing En2ERT2. En2ERT2 transfer was revealed by the detection of EnERT2 signal (green) in mCherry-positive cells (yellow cells, arrow). Scale bars: 20µm. (C,D) Internalisation of Eng2a and 2b in HEK293 cells. Following extracellular addition, internalisation of fluorescein-labelled En2, Eng2a or Eng2b protein (green) was visualised (C) and quantified (D) after 1h incubation at 37°C in the presence of 0.4% Trypan Blue (red) to quench extracellular fluorescence. MFI, mean fluorescence intensity. (E) Secretion of Eng2a and 2b. HEK293 cells expressing the indicated proteins under the control of doxycycline were cultured for 24h and cell surface accumulation of the secreted protein was monitored by flow cytometry. ***P<0.001. (F) Paracrine activity of Eng2a and Eng2b proteins. mRNA encoding En2ERT2, Eng2aERT2 or Eng2bERT2 were injected at the one-cell stage, the protein was activated with CYC at 50% epiboly and eye defects were scored at 30hpf. The error bars represent statistical errors (F) or s.e.m. (D,E). |
|
Intercellular transfer of Eng2b but not 2a is involved in DMB positioning. (A,B) Co-detection of Eng2 proteins with FITC-labelled 4D9 and TRITC-labelled 4G11 antibodies by immunohistochemistry (A) or western blot analysis on HeLa cells (B). Using both techniques, 4D9 recognised Eng2a and Eng2b, whereas 4G11 recognised only Eng2a. (C-H) Paracrine activity of endogenous En2 proteins. Zebrafish embryos were injected in the intercellular space at blastula stage with anti-myc or anti-Engrailed (4G11 or 4D9) antibody. Mesencephalon length was quantified by pax6 (C,E,G) or wnt1 (D,F,H) in situ hybridisation. Representative in situ hybridisation staining of pax6 (dorsal view) (C) and wnt1 (lateral view) (D). Measurements were distributed into seven size classes (smallest size, class 1) and plotted as cumulative frequency index. 4D9 extracellular injection induced mesencephalon shortening (E,F) in a dose-dependent manner (supplementary material Fig. S4), which was the case for neither anti-myc (E-H) nor 4G11 injection (G,H). Co-injection of the 4D9 antibody with its epitope peptide (pep) significantly reduced this effect (E,F). ***P<0.0001, Mann–Whitney tests. (I) Inhibition of homeoprotein transfer rescued the phenotype of Eng2b activation but not of Eng2a. Zebrafish embryos were injected at the one-cell stage with the indicated mRNAs and injected again in the extracellular space at the blastula stage with 4D9 anti-En antibody. CYC was added at 50% epiboly in the water bath to activate the protein. Eye phenotypes were scored at 2dpf. The error bars represent statistical errors. (J) Transcriptional activity of En2 and the two zebrafish Eng2. The MAP1B:luciferase reporter plasmid was transfected into HeLa cells along with an empty vector (Ctrl) or together with the indicated constructs, and cells were analysed for luciferase activity after 24h. a.u., arbitrary units. **P<0.01, ***P<0.001. |
|
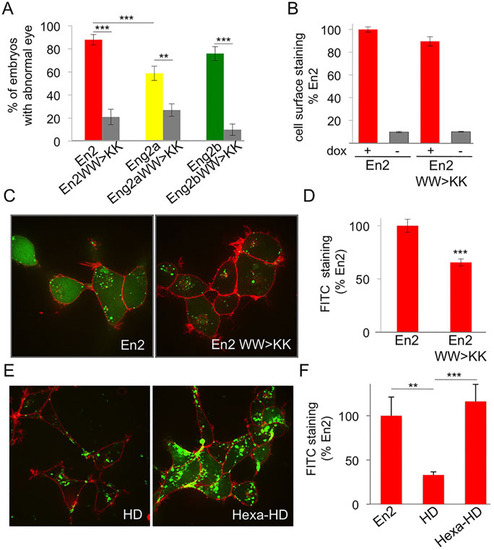
Pbx interaction domain controls En paracrine activity. (A) Phenotypic analysis of hexapeptide-mutated forms of En2 protein. Compared to their wild-type counterparts, the eye phenotype induced by the activation of hexapeptide mutants was drastically reduced. The error bars represent statistical errors. (B-D) Intercellular transfer of En2 hexapeptide mutant ex vivo. HEK293 cells expressing the indicated proteins under the control of doxycycline were cultured for 24h and cell surface accumulation of the secreted protein was monitored by flow cytometry (B). Internalisation of fluorescein-labelled En2, or En2 WW>KK (green) was visualised (C) and quantified (D) after 1h incubation at 37°C in the presence of 0.4% Trypan Blue (red) to quench extracellular fluorescence. (E,F) Internalisation of fluorescein-labelled En2, En2HD, Hexa-En2HD was visualised (E) and quantified (F) as described above. The error bars represent statistical errors (A) or the s.e.m. (B,D,F). **P<0.01, ***P<0.001. |