- Title
-
GSTT1 deletion is related to polycyclic aromatic hydrocarbons-induced DNA damage and lymphoma progression
- Authors
- Yang, F., Xiong, J., Jia, X.E., Gu, Z.H., Shi, J.Y., Zhao, Y., Li, J.M., Chen, S.J., Zhao, W.L.
- Source
- Full text @ PLoS One
|
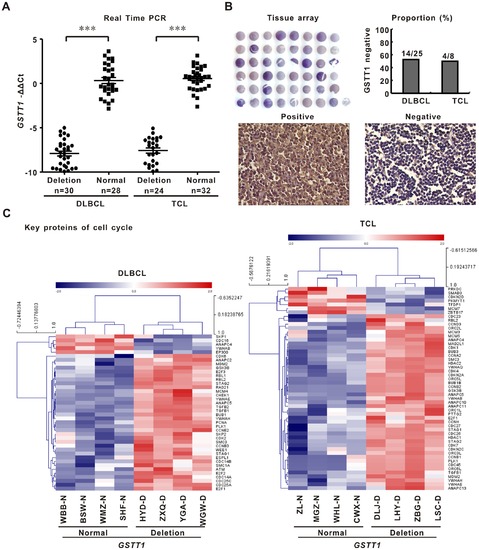
GSTT1 deletion is related to decreased gene and protein expression and enhanced cell cycle progression. A: GSTT1 gene expression assessed by quantitative real-time PCR in the GSTT1-deleting patients and normal controls. B: GSTT1 protein expression detected by tissue array. C: Geneset of cell cycle related proteins revealed by gene network and pathway analysis on microarray data of DLBCL and TCL. |
|
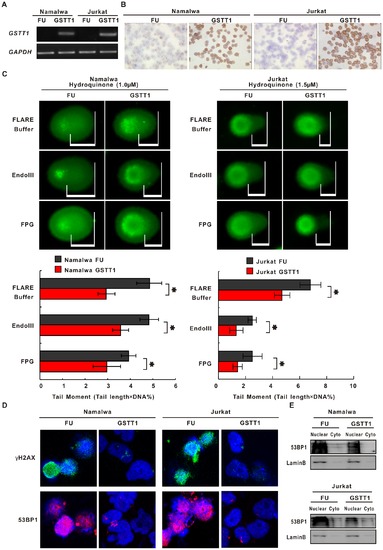
GSTT1 expression protects lymphoma cells from PAH-induced DNA damage. A: GSTT1 gene expression assessed by semi-quantitative PCR in Namalwa and Jurkat cells transfected with GSTT1 (GSTT1) and the negative control vector (FU). B: Images represent results from three independent experiments. GSTT1 protein expression detected by immunohistochemistry assay. C: DNA damage measured by alkaline and modified comet assay in Namalwa and Jurkat cells treated with Hydroquinone (Upper panels). Mean tail moments were calculated in the same cells (Lower panels). Data represents Mean ± SE from at least 50 cells in each group. D: Immunofluorescence assay of γH2AX and 53BP1 in Hydroquinone-treated lymphoma cells. *P<0.05 comparing with the FU cells. |
|
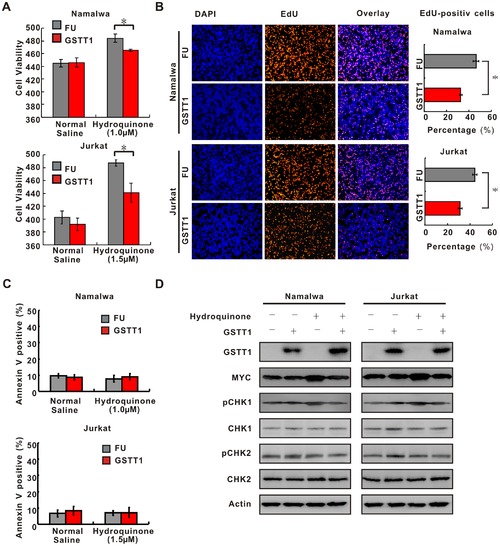
GSTT1 expression inhibits PAH-mediated lymphoma cell proliferation. A: Effect of GSTT1 expression on cell proliferation. 3×105 cells treated with normal saline or Hydroquinone were seeded and cell number was counted at 48 h by typan blue. Data represent Mean±S.E. of densitometric values from three individual experiments. B: EdU assay of Namalwa and Jurkat cells treated with normal saline or Hydroquinone. C: Cell apoptosis analyzed by flow cytometry. Histography indicates Mean±S.E. from three individual experiments. D: Key proteins of DNA damage and cell cycle detected by Western blot in Namalwa and Jurkat cells with or without Hydroquione treatment. *P<0.05 comparing with the FU cells. |
|
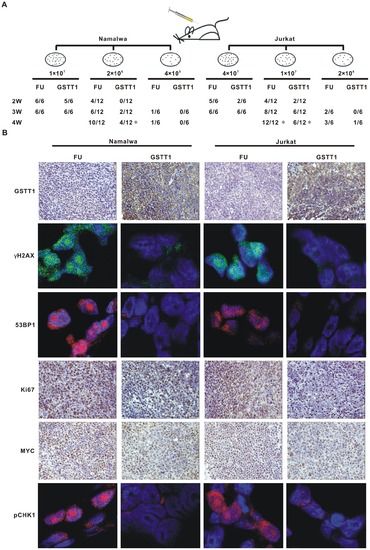
GSTT1 expression reduces tumorogeneity of PAH-treated lymphoma cells. A: Tumor formation in nude mice. Indicated amount of Hydroquinone treated cells were injected subcutaneously and tumorigenicity was reported as numbers of tumors formed per numbers of mice injected. B: Expression of GSTT1, Ki67 and MYC detected in tumor tissues by immunohistochemistry assay, as well as pCHK1, γH2AX and 53 BP1 by immunofluorescence assay. *P<0.05 comparing with the FU cells. |
|
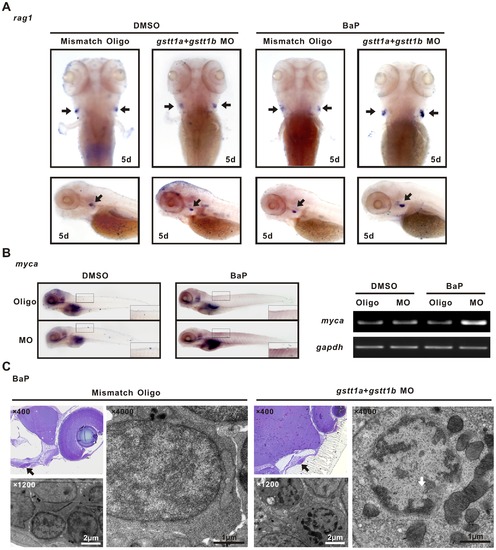
Knock-down of gstt1a and gstt1b promotes lymphocyte proliferation exposed to BaP. A; WISH images showed the rag1 expression in the thymus (arrows) of differently treated 5 dpf embryos. B: In situ analysis of myca at 5 dpf. The morphants showed increased expression of myca in microinjected gstt1a and gstt1b morpholino exposed to BaP (Left panels), semi-quantitative PCR showed similar expression pattern in embyos (Right panels). C: Ultrastructure of thymic lymphocytes from 5 dpf larvae exposed to BaP. Images represent results from three independent experiments and each group contains 30 morphants. EXPRESSION / LABELING:
|
|
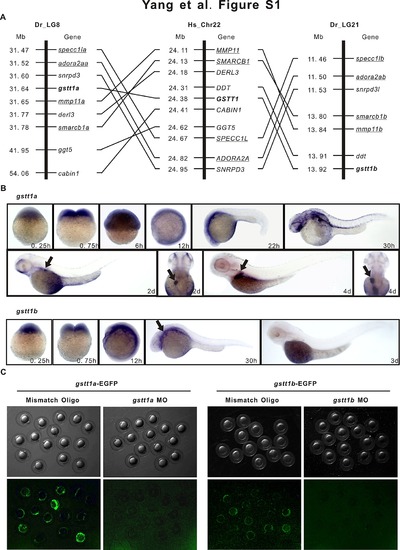
GSTT1 is evolutionarily conserved and expressed ubiquitously during embryonic development. A: Comparison of the syntenic relationship of the zebrafish gstt1 genes with the human orthologue. Orthologous gstt1 genes symbols were in bold. Other pairs of duplicated genes (e.g. mmp11a and mmp11b) on zebrafish and the human (e.g. MMP11) orthologue are underlined. Hs_Chr, Homo sapiens chromosome, Dr_LG, Danio rerio linkage group, Mb, megabase. B: Expression of zebrafish gstt1a and gstt1b in wild-type AB strain embryo during embryonic development. C: Efficiency validation of EGFP reporter expression by gstt1a morpholino and gstt1b morpholino. Images represent the typical outcome of three independent experiments and each group contains 30 morphants. |