- Title
-
A Polymorphism in IRF4 Affects Human Pigmentation through a Tyrosinase-Dependent MITF/TFAP2A Pathway
- Authors
- Praetorius, C., Grill, C., Stacey, S.N., Metcalf, A.M., Gorkin, D.U., Robinson, K.C., Van Otterloo, E., Kim, R.S., Bergsteinsdottir, K., Ogmundsdottir, M.H., Magnusdottir, E., Mishra, P.J., Davis, S.R., Guo, T., Zaidi, M.R., Helgason, A.S., Sigurdsson, M.I., Meltzer, P.S., Merlino, G., Petit, V., Larue, L., Loftus, S.K., Adams, D.R., Sobhiafshar, U., Emre, N.C., Pavan, W.J., Cornell, R., Smith, A.G., McCallion, A.S., Fisher, D.E., Stefansson, K., Sturm, R.A., and Steingrimsson, E.
- Source
- Full text @ Cell
|
Intron 4 of IRF4 Drives Expression in Melanocytes (A and B) Results of the luciferase assays in melan-Ink4a-Arf and SK-MEL-28 cells are shown. Pr, promoter; Luc, luciferase reporter gene; IRF4 enh, 450 bp fragment from the fourth intron of IRF4 containing rs12203592. “IRF4 enh” fragments are identical except for the base at position rs12203592 as indicated (“C” is the ancestral allele; “T” is the derived allele). The x axis shows fold luciferase activity relative to the minimal promoter alone, which is normalized to one (1). p values are calculated by Kolmogorov-Smirnov test. The result is also highly significant by other nonparametric tests (p = 0.007937 by Wilcoxon rank sum test for both melan-Ink4a-Arf and SK-MEL-28) and by the standard parametric t test (p = 0.001686 for melan-Ink4a-Arf; p = 0.01032 for SK-MEL-28). Each bar represents the average of five biological replicates per reporter construct. Error bars represent SD. (C) Reporter constructs contain either the rs12203592-C version of the IRF4 intron 4 element (IRF4) or the rs12203592-T version (IRF4snp) upstream of the GFP reporter after injection into zebrafish. The numbers represent the number of melanophores (blue) and embryos (red) seen to be positive (pos.) for GFP. The difference between the melanophores containing the wild-type allele of intron 4 of IRF4 compared to the mutant allele (rs12203592) is statistically significant (p = 0.0023, unpaired t test). (D) Image shows a GFP-positive zebrafish embryo with a GFP-positive melanophore in the inset. |
|
MITF Regulates Expression of IRF4 (A) The expression of the Mitf and Irf4 mRNAs is reduced in hearts from Mitfmi-vga9 mutant mice, as determined by qPCR analysis. Data are represented as mean ± SEM. (B) Expression of the MITF, IRF4, TFAP2a, DCT, and TYR genes upon treatment with shRNA against MITF, IRF4, TFAP2A, and negative control in 501mel melanoma cells as determined by qPCR analysis is shown. Data are represented as mean ± SEM. (C) Western blot shows expression of the MITF, TFAP2A, IRF4, and β-actin proteins in 501mel cells after shRNA treatment. Intensity quantification is relative to actin-loading control. See also Figure S2. (D) Overexpression of the mouse Mitf (mMitf) cDNA (expressed from pcDNA3.1) in 501mel melanoma cells and in HEK293T embryonic kidney cells affects expression of the TYR and IRF4 mRNAs as assayed by RT-qPCR, whereas β-actin expression is unchanged. mMitf was detected with species-specific primers that only recognize the mouse Mitf gene. See also Figure S3. See also Figure S2 and Table S3. |
|
MITF and TFAP2A Affect IRF4 Expression by Binding Regulatory Elements in Intron 4 (A) Schematic view presents the IRF4 gene. The sequence shows a comparison of MITF- and TFAP2A-binding sites in intron 4 of IRF4 among humans (H sap [Homo sapiens]), gorilla (G gor [Gorilla gorilla]), chimpanzee (P tro [Pan troglodytes]), and mouse (M mus [Mus musculus]). The location of the rs12203592 polymorphism is indicated. (B) ChIP analysis of a GST-tagged MITF protein (a-GST AB) was performed in 501mel cells. Primers specific for HSPA1B (negative control), TYR (positive control), and intron 4 of IRF4 are indicated. The GST-tagged Mitf only precipitated TYR and IRF4 sequences. (C) ChIP analysis of TFAP2A in 501mel cells is presented. PCR products specific for TGFA (positive control), HSPA1B (negative control), and intron 4 of IRF4 are shown. TFAP2A only precipitated TGFA and INT4 sequences. (D) Gel shift analysis shows the binding of TFAP2A to the ancestral sequence (INT4-WT) in intron 4 of IRF4 (coordinates chr6: 396309–396333 in build GRCh37.p5 of the human genome), but not to the rs12203592-T sequence (INT4-RS) or to a completely mutated sequence (INT4-mut). (E) Luciferase reporter assays were performed in 501mel melanoma cells using intron 4 of IRF4 as a reporter. The ancestral IRF4 intron 4 sequence (intron 4, blue), the rs12203592-T polymorphic sequence (intron 4-rs), a sequence where all MITF-binding sites were mutated (intron 4-3xE), and a sequence containing mutated MITF sites and the rs12203592-T polymorphism (intron 4-3xE+rs) were tested. The luciferase reporters were cotransfected with the wild-type MITF and TFAP2A proteins and with a dominant-negative version of MITF (MITFmi). Statistical analysis was done using unpaired t test. Data are represented as mean ± SEM. (F) Cells homozygous for the ancestral allele (CC) express significantly higher levels of IRF4 than cells homozygous for the rs12203592-T polymorphism (TT). Fold expression data are represented relative to pooled mean TT expression ± SEM. Statistical analysis was performed using ANOVA (p < 0.001). (G) A western blot shows expression of the IRF4 protein in melanocytes from CC and TT individuals. Intensity quantification is relative to GAPDH-loading control. Note the decrease of basal IRF4 protein in the TT melanocyte cell lines. See also Figures S3, S4, and S5. |
|
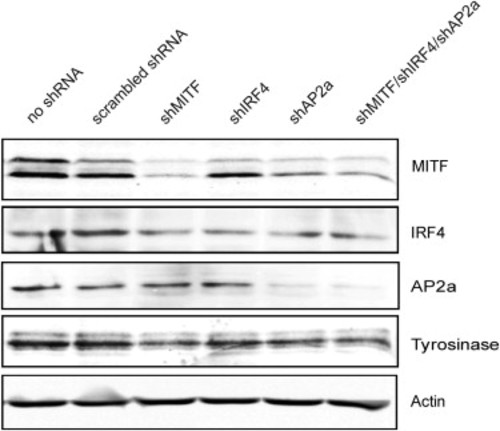
Western Analysis Showing MITF, IRF4, TFAP2A, and TYR Expression upon shRNA Treatment, Related to Figure 4C Western blot showing expression of the MITF (Thermo Fisher C5), TFAP2A (Abcam ab52222), IRF4 (Cell Signaling, IRF4 (CST #4299), TYR (Santa Cruz sc-7833) and β-actin (Millipore C4 MAB1501R) proteins in 501mel cells after shRNA treatment. |
Reprinted from Cell, 155(5), Praetorius, C., Grill, C., Stacey, S.N., Metcalf, A.M., Gorkin, D.U., Robinson, K.C., Van Otterloo, E., Kim, R.S., Bergsteinsdottir, K., Ogmundsdottir, M.H., Magnusdottir, E., Mishra, P.J., Davis, S.R., Guo, T., Zaidi, M.R., Helgason, A.S., Sigurdsson, M.I., Meltzer, P.S., Merlino, G., Petit, V., Larue, L., Loftus, S.K., Adams, D.R., Sobhiafshar, U., Emre, N.C., Pavan, W.J., Cornell, R., Smith, A.G., McCallion, A.S., Fisher, D.E., Stefansson, K., Sturm, R.A., and Steingrimsson, E., A Polymorphism in IRF4 Affects Human Pigmentation through a Tyrosinase-Dependent MITF/TFAP2A Pathway, 1022-1033, Copyright (2013) with permission from Elsevier. Full text @ Cell