- Title
-
Fgf differentially controls cross-antagonism between cardiac and haemangioblast regulators
- Authors
- Simões, F.C., Peterkin, T., and Patient, R.
- Source
- Full text @ Development
|
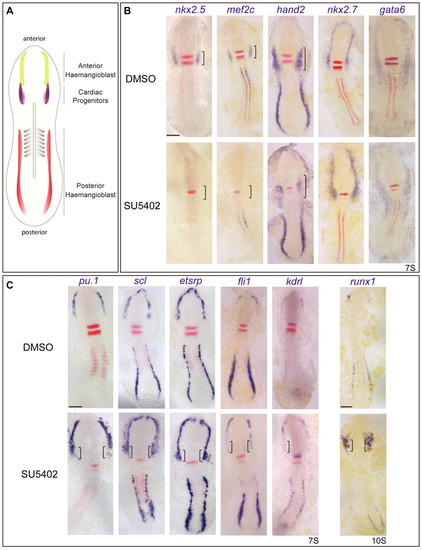
Fgf signalling inhibits blood and endothelial gene expression in the heart field. (A) Schematic highlighting the anterior blood/endothelial precursor or haemangioblast population (yellow) rostral to cardiac precursors (purple) in 7-somite zebrafish embryos. Another haemangioblast population is found in the posterior lateral plate mesoderm (red). (B) Expression of the cardiac markers (brackets) nkx2.5, mef2c and hand2 is severely reduced in embryos treated with the Fgf inhibitor SU5402 compared with control DMSO-treated siblings. A few hand2+ cells remain in the cardiac mesoderm of inhibitor-treated embryos. Anterior mesoderm specification is unaffected as revealed by normal expression of the anterior lateral plate mesoderm (ALPM) markers nkx2.7 and gata6. (C) Blood and endothelial gene expression is upregulated and ectopic (brackets) in the ALPM of SU5402-treated embryos. Images show flat-mounted embryos, anterior to the top. Somites were stained with myoD or uncx4 (red) for staging. Rhombomeres 3 and 5 were stained with krox20 (red stripes) as a positional marker. Reduction in rhombomere 5 reveals that Fgf signalling is reduced, together with short body axis and posterior mesoderm defects. Scale bars: 100 μm. |
|
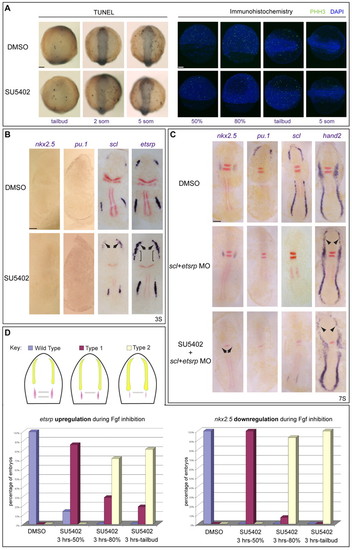
Fgf induces cardiac specification independently of, but concomitant with, haemangioblast repression. (A) Neither apoptosis nor proliferation is increased in SU5402-treated zebrafish embryos. Neither TUNEL nor phospho-histone-H3 (PHH3, green) staining increased in Fgf-depleted embryos during gastrulation or at the 5-somite (5 som) stage. Nuclei are stained with DAPI (blue). (B) Fgf inhibition of haemangioblast gene expression precedes nkx2.5 expression. When Fgf was reduced, the haemangioblast programme was upregulated in the anterior lateral plate mesoderm (ALPM) before the onset of nkx2.5 expression. Embryos were treated from 3 hpf with SU5402 and RNA expression was assayed at the 3-somite stage. No differences were observed for nkx2.5 or pu.1, compared with DMSO-treated embryos, but scl and etsrp were upregulated in the ALPM (arrows). etsrp was also ectopically expressed in adjacent mesoderm (brackets). (C) scl and etsrp morpholino (MO) co-injection led to severe downregulation of anterior haemangioblast gene expression (pu.1 and scl) with associated rostral expansion of cardiac hand2 expression (arrowheads). nkx2.5 expression was unchanged at this stage but, when Fgf signals were also deficient in scl/etsrp double morphants (SU5402 + scl+etsrp MO), nkx2.5 expression was absent (arrows). Downregulation of hand2 expression in the cardiac region was also seen; however, rostral expression was still observed (arrowheads). (D) Fgf signalling is required concomitantly for the establishment of cardiac and haemangioblast fates. Fgf was inhibited for different periods of time and expression of etsrp and nkx2.5 assessed by in situ hybridisation at the 7-somite stage. Phenotypes were categorised as wild type (blue), type 1 (purple) or type 2 (yellow) (see inset key diagram), where type 1 represents downregulation of nkx2.5 with associated expansion of etsrp, and type 2 embryos show strong downregulation or absence of nkx2.5 and coupled expansion and upregulation of etsrp. Percentages of embryos showing increased etsrp or decreased nkx2.5 expression are shown. The experiment was repeated three times for 10 μM treatments and a representative experiment is shown, n>70 for each experimental point. Images show flat-mounted embryos, anterior to the top. Double-staining for myoD and krox20 (red) was used for staging and orientation. Scale bars: 100 μm. |
|
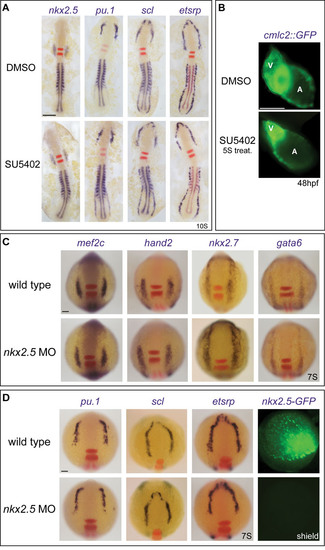
The cardiac and blood/endothelial programmes are cross-antagonistic. (A) Nkx2.5 RNA over-expression (50 pg) from the 1- to 2-cell stage reduced etsrp, scl and pu.1 and expanded hand2 expression in the anterior lateral plate mesoderm (ALPM; arrowheads) at the 7-somite stage. The anterior mesodermal marker gata6 was unaffected in the same cells. Nkx2.5-GFP fusion expression was visualised at the shield stage to confirm injection and expression. Dorsal views of 7-somite whole zebrafish embryos are shown, anterior to the top; for fluorescence, lateral views of shield stage whole embryos are shown, dorsal to the right. (B) An increase in endothelium and myelopoiesis was evident throughout development upon depletion of Fgf. Embryos treated with SU5402 at 3 hpf were assessed for endocardium by expression of kdrl and cdh5 at the 16-somite stage, whereas pu.1 was assessed for myeloid development. Increased endocardial (arrows) and myeloid gene expression was observed. (C) Similar treatment increased kdrl::GFP+ endothelial cell numbers, with concomitant loss of myocardium at 36 hpf (white arrow; stained with MF20 antibody). (D) Increased differentiated myeloid cells (l-plastin+) seen at 30 hpf. (E) Nkx2.5-induced downregulation of the haemangioblast programme is not sustained throughout development. Myeloid gene expression (pu.1) was downregulated at the 16-somite stage upon nkx2.5 over-expression, and at 26 hpf l-plastin remained depleted. However, expression of cmlc2, kdrl and cdh5 appeared similar to wild type at 26 hpf. Dorsal views are shown, anterior to the top, except in panel D and 26 hpf embryos in panel E in which lateral views are shown, anterior to the left. Scale bars: 100 μm. |
|
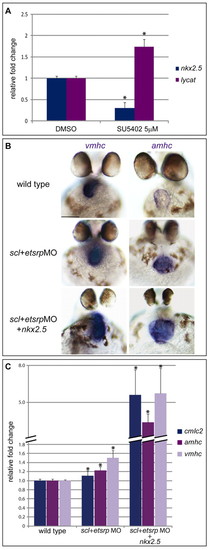
Fgf is working via nkx2.5 and lycat during gastrulation to establish cardiac and haemagioblast fates. (A) Quantitative PCR on cDNA obtained from RNA extracted from 50% epiboly zebrafish embryos treated with DMSO and SU5402, assayed for lycat and nkx2.5 expression normalised to gapdh. Shown are fold changes in expression, relative to the DMSO control, from three biological replicates. Error bars indicate s.e.m., asterisks indicate statistically significant differences (P<0.001) compared with DMSO controls. (B,C) The scl and etsrp morpholino combination increased amhc and vmhc expression, as seen by in situ hybridisation (B) and qPCR (C; P<0.02) at 48 hpf. However, expression was substantially more upregulated in scl+etsrpMO and nkx2.5 RNA embryos (P<0.05). A similar trend was observed for cmlc2 (P<0.05). Three biological replicates were analysed. Error bars indicate s.e.m., asterisks indicate statistically significant differences compared with wild type and scl+etsrpMO siblings. Scale bars: 100 μm. |
|
Fgf inhibition in zebrafish. (A) The Fgf target gene pea3 is downregulated at the 20-somite stage upon treatment with SU5402 from 3 hpf. Dorsal and lateral views rae shown, with anterior to the right and left, respectively. (B) Quantitative PCR on cDNA obtained from RNA extracted from 7-somite embryos treated with DMSO and SU5402, and assayed for nkx2.5, pu.1 and gata6 expression normalized to gapdh. Shown are fold changes in expression relative to the DMSO control, from one representative experiment. Three biological replicates were analysed, each time in triplicate. Error bars indicate s.e.m., asterisks indicate statistically significant differences (P<0.05) compared with DMSO controls. (C,D) fgf3 and fgf8 combine to specify heart versus haemangioblast cell identity. Cardiac (C) and blood/endothelial (D) gene expression effects at the 7-somite stage, when fgf3 and fgf8 were knocked down individually (fgf3 MO, fgf8 MO) or in combination (fgf3+8 MO) are shown. Percentage of embryos with downregulation of cardiac markers (red arrowheads) or upregulation (black arrowheads) plus or minus expansion (brackets) of anterior haemangioblast gene expression are shown. Embryos are flat mounted, anterior to the top. krox20 (red) acted as a positional marker and revealed the loss of rhombomere 5 expression when both fgf3+8 expression was inhibited. Scale bars: 100 μm. |
|
Fgf signalling affects the relative size of the haemangioblast and cardiac populations during a limited time window. (A) SU5402 inhibition performed at the 5-somite stage, after specification of cardiac and blood/endothelial cells, can no longer affect their identity. No difference in expression of the cardiac marker nkx2.5 or the haemangioblast genes pu.1, scl and etsrp was observed at the 10-somite stage, assayed by in situ hybridization, compared with DMSO siblings. Embryos are flat mounted, anterior to the top. Double staining for krox20 (red) provided a landmark. Note that rhombomere 5 expression is no longer reduced, in accordance with observations by Walshe et al. (Walshe et al., 2002). myoD (blue in nkx2.5-, pu.1- and scl-stained embryos, red in etsrp-stained embryos) was used for staging. (B) The overall heart size was reduced on depletion of Fgf signalling from 3 hpf treatment, as seen in cmlc2::GFP transgenic fish at 48 hpf (not shown) but, when Fgf signalling was inhibited from 5S (5 somite stage) onwards, the development of the atrial chamber was no longer affected. A, atrium; V, ventricle. (C,D) nkx2.5 is not essential for anterior lateral plate mesoderm (ALPM) specification. Morpholino (MO) knock-down of nkx2.5 had no effect on cardiac specification (C) or anterior haemangioblast gene expression (D) at the 7-somite stage. nkx2.5-GFP fusion expression was completely abolished at shield stage, confirming MO efficiency. Scale bars: 100μm. |
|
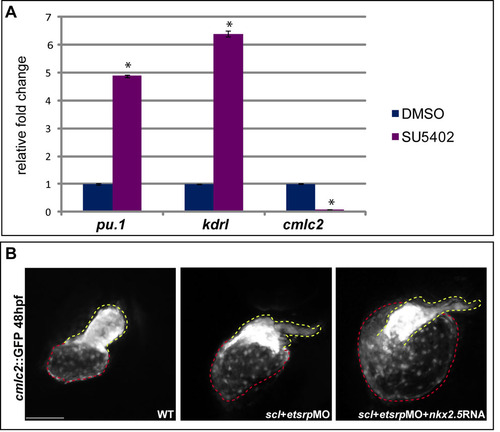
Reprogramming of cardiac and haemangioblast cells by Fgf signalling is mediated by lycat and nkx2.5. (A) Quantitative PCR on cDNA obtained from RNA extracted from the heart region of 36 hpf DMSO- and SU5402-treated embryos assayed for pu.1, kdrl and cmlc2 expression normalized to gapdh. Shown are fold changes in expression relative to the DMSO control, from one representative experiment. Four biological replicates were analysed in triplicate. Error bars indicate s.e.m., asterisks indicate statistically significant differences (P<0.0005) compared with DMSO controls. (B) cmlc2::GFP transgenic fish at 48 hpf show an overall heart size increase when injected with scl+etsrpMO. The increase is even greater in scl+etsrp morphants that have been co-injected with nkx2.5 mRNA. Dashed lines outline the atrium (red) and the ventricle (yellow). Scale bar: 100μm. |