- Title
-
Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration
- Authors
- Hoptak-Solga, A.D., Nielsen, S., Jain, I., Thummel, R., Hyde, D.R., and Iovine, M.K.
- Source
- Full text @ Dev. Biol.
|
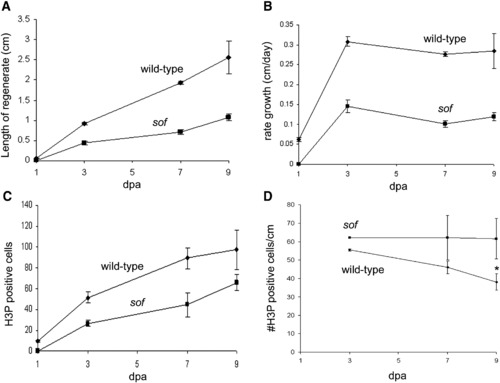
Growth rate and the number of mitotic cells are reduced in sofb123 mutants during regeneration. Measurements of regenerates in wild-type and sofb123 fins were conducted over a period of 9 days. (A) The length of the sofb123 regenerate is half as long as the wild-type regenerate. Standard deviation is shown. (B) The rate of regeneration is slower in sofb123 mutants. Standard deviation is shown. (C) The number of mitotic (H3P positive) cells is reduced in sofb123 mutants compared to wild-type. Standard deviation was calculated and the Student's t-test reveals all data points between wild-type and sofb123 are statistically different (p = 0.0001; 0.0008; 0.0042; 0.0454 for 1, 3, 7, 9 dpa). (D) The number of mitotic cells per cm is the same between wild-type and sofb123 fins. Standard deviation was calculated and the Student's t-test reveals data points through 7 dpa are not statistically different (p > 0.06). The data points at 9 dpa are statistically different, p = 0.0001 (*). PHENOTYPE:
|
|
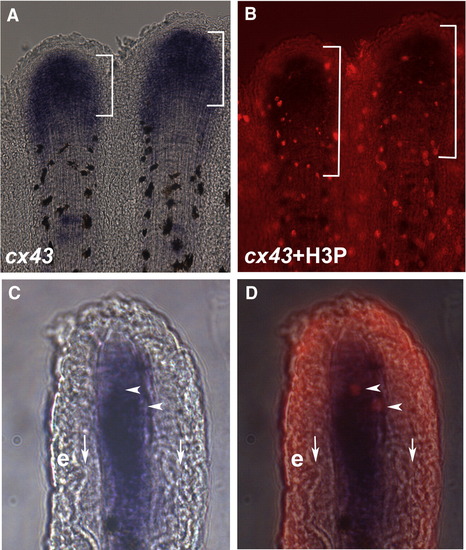
cx43 is expressed in mitotic cells. Regenerating fins were labeled for both cx43 mRNA and H3P. (A) Whole mount in situ hybridization shows cx43 expression (brackets). The cx43 positive compartment measured from 70–140 μm from the distal end of the fin. (B) Double-staining for H3P and cx43 in situ hybridization. A distance of 250 μm from the distal end of the fin is shown in brackets. (C) Brightfield image of a single cryosection showing cx43 expression in the distal mesenchymal compartment. (D) Brightfield plus fluorescence reveals that the two H3P positive cells in this section are coincident with cx43 positive cells. Vertical arrows point to bone matrix; e, epithelium; arrowheads point to doubly-labeled cells. EXPRESSION / LABELING:
|
|
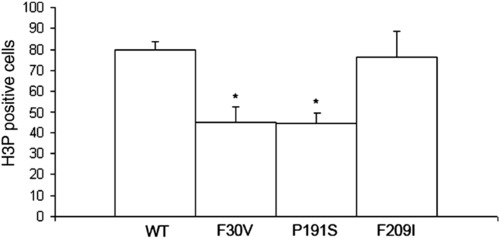
Cell proliferation is reduced in missense alleles of cx43. Fins were harvested at 3 dpa and stained for H3P detection. A statistically significant reduction in the number of H3P cells was observed for both the Cx43-F30V (p = 0.0001) and the Cx43-P191S (p = 0.0002) alleles in comparison to wild-type. The weaker Cx43-F209I allele was not statistically different from wild-type (p = 0.638). Standard deviation is shown for all alleles. PHENOTYPE:
|
|
Cx43 protein expression is reduced in sofb123 mutants. (A) An antibody raised against zebrafish Cx43 detects a GST fusion protein with the carboxy tail of Cx43 (GST-Cx43CT). Increasing amounts of E. coli lysates expressing GST-Cx43CT were loaded and tested for antibody detection (left). When the antibody was pre-incubated with the Cx43 target sequence, binding to the immunoblot was competed. (B) Protein levels of Cx43 in sofb123 regenerating fins and whole body are reduced by greater than 70%. Tubulin was used as a loading control and to normalize the loading of fin or whole body lysates. (C) Percent reductions of anti-Cx43 antibody binding for the antibody competition (A) and sofb123 Cx43 levels (B). (D) Cx43 expression in wild-type fins. Distal tip staining in wild-type (E) and sofb123 fins (F). (G) Cx43 expression is substantially reduced in sofb123 mutant fins. Arrows point to Cx43 staining at the joints. EXPRESSION / LABELING:
PHENOTYPE:
|
|
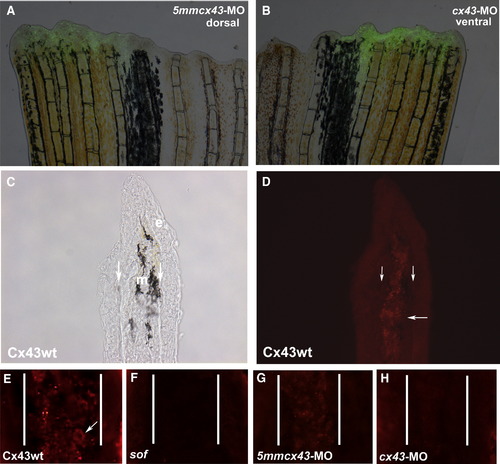
Injection and electroporation of cx43 morpholinos confirms targeted knockdown of Cx43. (A,B) Fins were injected with either 1.2 mM 5mmcx43-MO or 1.2 mM cx43-MO and immediately electroporated. Cellular uptake is shown by fluorescein fluorescence. (C,D) Cryosections through fins stained with Cx43 antibody show mesenchymal staining in wild-type fins. Brightfield and fluorescence are shown. Vertical arrows indicate bone matrix, horizontal arrow points to one area of punctate staining. (E) Closer examination of Cx43 staining reveals punctate plasma membrane staining. Arrow points to one cell. (F) sofb123 regenerates exhibit a reduction in Cx43 protein in the mesenchyme (G) Fins injected/electroporated with the 5mmcx43-MO show similar expression of Cx43 as wild-type fins. (H) Fins injected/electroporated with the targeting morpholino cx43-MO exhibit a reduction in Cx43 expression. Vertical lines in panels E–H distinguish epithelium and mesenchymal compartments. EXPRESSION / LABELING:
|
|
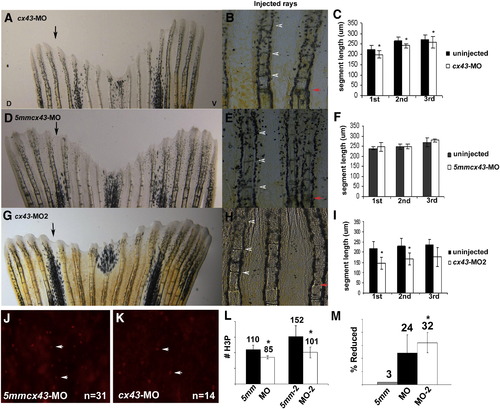
Knockdown of Cx43 recapitulates the sof phenotype. Wild-type fins were injected/electroporated in dorsal fin rays with either targeting morpholino (cx43-MO, cx43-MO2) and the respective control morpholino. The ventral side of the fin was electroporated only. At 4 dpe, segment length was measured for the first three newly formed segments for each fin ray. (A) Fin electroporated with 1.2 mM cx43-MO. (B) Segment size was measured for both injected and uninjected fin rays. (C) Graph comparing segment length (μm) for the cx43-MO and the uninjected rays. There is a significant difference in segment length for all three segments (p < 0.01). (D) Fin electroporated with 1.2 mM 5mmcx43-MO. (E) Segment size was measured for both injected and uninjected fin rays. (F) Graph comparing segment length (μm) for the 5mmcx43-MO and the uninjected rays. There is no significant different in segment length (p = 0.25, p = 0.94, p = 0.08, respectively). (G) Fin electroporated with 1.2 mM cx43-MO2. (H) Segment size was measured for both injected and uninjected fin rays. (I) Graph comparing segment length (μm) for the cx43-MO2 and the uninjected rays. There is a significant difference in segment length for all three segments (p < 0.01). Black arrows indicate the injected lobe, red arrows indicate the amputation plane, arrowheads designate newly formed joints. (J–L) Wild-type fins were injected/electroporated with either targeting or control morpholinos, harvested at 1 dpe and processed for H3P detection. Representative images of H3P positive cells are shown in panels J, K. (J) Dorsal rays were injected/electroporated with 5mmcx43-MO. (K) Ventral fin rays were injected/electroporated cx43-MO. (L) A significant reduction in H3P number was observed for either targeting morpholino (cx43-MO, p < 0.01 and cx43-MO2, p = 0.01) when compared to their corresponding controls. (M) The cx43-MO2 appears more effective at cx43 gene knockdown than cx43-MO since the percentage of H3P positive cells was more greatly affected when the cx43-MO2 was used (p < 0.05 when comparing the percent reduction of H3P positive cells of cx43-MO and cx43-MO2). |
Reprinted from Developmental Biology, 317(2), Hoptak-Solga, A.D., Nielsen, S., Jain, I., Thummel, R., Hyde, D.R., and Iovine, M.K., Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration, 541-548, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.