- Title
-
The Zebrafish Progranulin Gene Family and Antisense Transcripts
- Authors
- Cadieux, B., Chitramuthu, B.P., Baranowski, D., and Bennett, H.P.
- Source
- Full text @ BMC Genomics
|
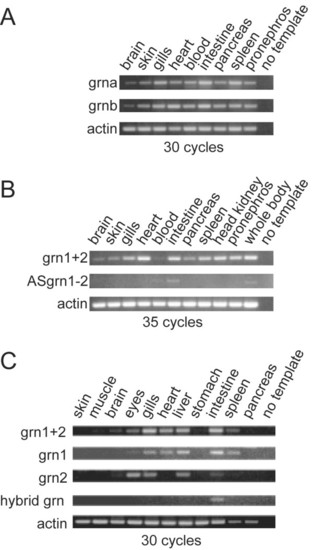
RT-PCR Analysis of zebrafish grns in adult tissues. Panel A: Zebrafish grna and grnb are ubiquitously expressed in various organs. Panel B: A comparison of the combined expression of grn1 and grn2 (grn1+2) relative to their antisense transcript. An increased number of cycles was used in the PCR to allow for the detection of ASgrn1-2 transcripts. Panel C: grn1, grn2 and hybrid grn are differentially regulated with the latter expressed only in the intestine. Results shown were generated using the forward and reverse1 primer pair (Additional File 10). Identical results were obtained using the forward and reverse2 primer pair (not shown). Hybrid grn was amplified using a grn1 forward and grn2 reverse primer combination. No product was obtained using a grn2 forward and grn1 reverse primer pair (not shown). Number of cycles for each reaction is indicated. Amplified PCR products were analyzed by electrophoresis next to a 100-bp DNA ladder. No template and amplification of actin mRNA were used as negative and positive controls, respectively. Similar results were obtained with two other experiments. EXPRESSION / LABELING:
|
|
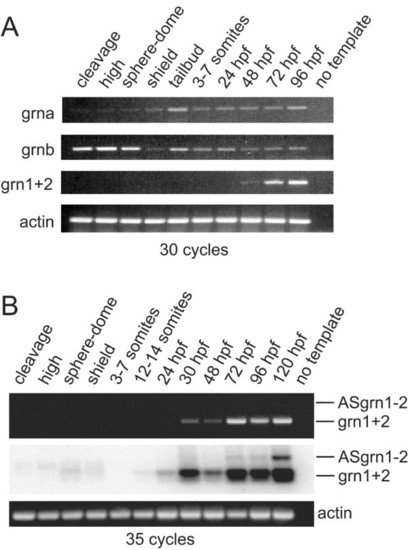
RT-PCR analysis of zebrafish grn expression during development. Panel A: grna and grnb transcripts are detected throughout all stages of development, whereas grn1 and grn2 expression is first detected by 48 hours post-fertilization. Maternal expression of grnb is more abundant than grna. Panel B: Combined expression of grn1 and grn2 relative to their antisense transcript. Ethidium bromide stain reveals the presence of sense transcription only (top). Detection of the antisense transcript is revealed by using a 32P-labelled oligo as probe that recognizes both sense and antisense amplicons after Southern transfer. Note weak expression of grn1 and/or grn2 at earlier stages of development. Numbers of cycles used for the PCR are indicated. No template and actin were used as negative and positive controls, respectively. Developmental stages are as follows according to Kimmel et al. 1995: cleavage (16-cell); high (mid-blastula, 3 hpf); sphere-dome (late blastula, 4-4.3 hpf); shield (50% epiboly, 6 hpf); tailbud (10 hpf); 3-7 somites (11-12 hpf); 12-14 somites (14-16 hpf); early (24 hpf) and late (48 hpf) pharyngula; hatching (72 hpf); ealy larval period (96 and 120 hpf). Gene specific primers and amplicon sizes are listed in Additional File 10. |
|
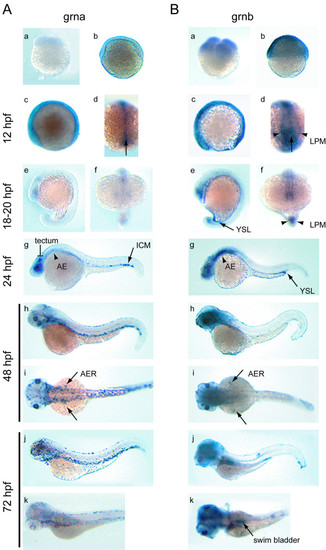
Developmental expression analysis of zebrafish grna and grnb mRNAs by whole mount in situ hybridization. An ontogeny of expression conducted for grna (A) and grnb (B) revealed similar expression patterns for these genes from fertilization to late segmentation stage (a-f), with grna being weaker than grnb. At the 4-cell stage (a) and 50% epiboly (b) ubiquitous expression is observed for grnb only. A lateral view of the 6-somite stage embryo (c) reveals discernable ubiquitous expression above background levels for grna, and increased grnb expression in the epithelium of the eye primordium and CNS, as well as the caudal region. In a dorsal view of the same animals (d), caudal expression in the axial mesoderm (arrow) is observed for both genes, whereas only grnb is detected in the paraxial mesoderm (arrowheads). Lateral (e) and frontal (f) views of the late somitogenesis stage embryo (18-20 hpf) show continued expression in the eye primordium, CNS and tailbud for both genes. In addition, grnb can be detected in the YSL (arrow) (e) and the adaxial cells (arrowheads) (f) flanking the axial mesoderm. At 24 hpf (g), grna expression is found in the tectum and eye retina, in a diffuse pattern in the anterior endoderm (arrowhead) and in a punctuate pattern within the ventral tail region of the ICM (arrow), whereas elevated expression persists for grnb in the forebrain, midbrain and ventral hindbrain region, the eyes, as well as in the YSL, concentrated at the tip of the yolk extension (arrow). In a lateral view at 48 hpf (h), grna expression in the ICM extends rostrally, is detected in the head vasculature, and is now apparent in the skin epithelium, whereas in a lateral view (i) both grna and grnb are transiently expressed in the AER of the pectoral fin buds (arrows). At 72 hpf (j), grna, but not grnb, is expressed in presumed dispersed leukocytes, while in a dorsolateral view (k), grnb can be detected in the swim bladder (arrow). AE, anterior endoderm; AER, apical ectodermal ridge of the pectoral fin buds; ICM, intermediate cell mass; LPM. lateral plate mesoderm; YSL, yolk syncytial layer. EXPRESSION / LABELING:
|
|
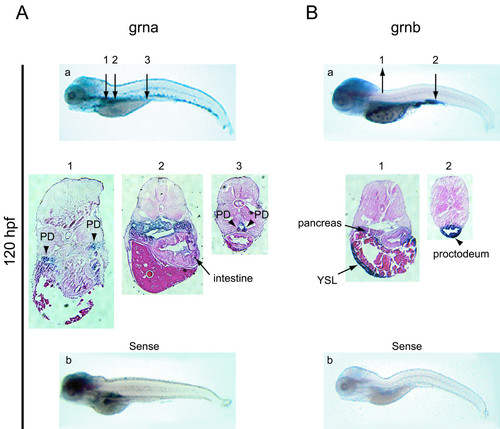
Expression analysis of zebrafish grna and grnb mRNAs by whole mount in situ hybridisation at 5 dpf. At 120 hpf grna (A) exhibits widespread expression in the visceral region, including the pronephric kidneys (arrowheads in sections 1 and 3) and intestine (arrow in section 2), while grnb (B) remains expressed in the YSL and pancreas (arrows in section 1) and is strong in the proctodeum region of the intestine (arrow in section 2). At this stage, a hybridization signal for the sense riboprobe to grna, but not grnb, is detected in the brain, intestine and pronephric ducts (b). Numbered arrows denote the position of corresponding sections shown below (magnified 10x). PD, pronephric ducts; YSL, yolk syncytial layer. |
|
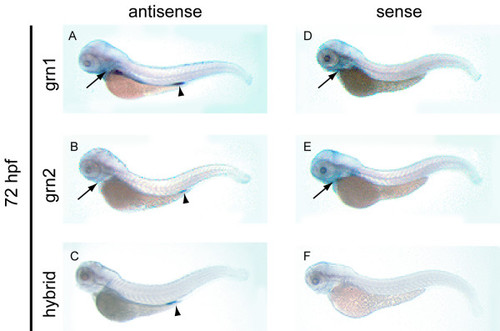
Expression analysis of grn1, grn2, hybrid grn, and ASgrn1-2 in the hatching stage zebrafish embryo by whole mount in situ hybridization. Panel A: grn1 is expressed in the intestine and pharyngeal region (arrow), and at low levels in the pronephric ducts. Panel B: In contrast, grn2 mRNA is only weakly detected in the pharyngeal region (arrow) and the proctodeum (arrowhead), and is occasionally found in dispersed leukocytes (not shown). Panel C: The abundance of the trans-spliced product (hybrid grn) is stronger than grn2 in the proctodeum (arrowhead), but absent in the pharyngeal region. Panels D-F: The corresponding sense riboprobes to grn1 and grn2, but not to hybrid grn, detect ASgrn1-2 expression in the pharyngeal region (arrows). These expression patterns were reproduced in at least three independent experiments. EXPRESSION / LABELING:
|
|
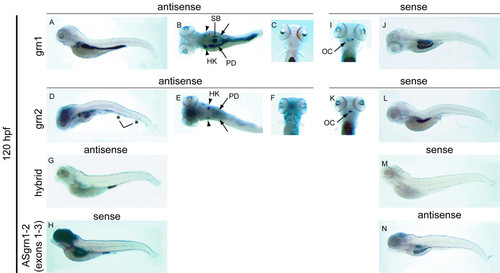
Expression analysis of grn1, grn2, hybrid grn, and ASgrn1-2 in the 5 day-old zebrafish larva by mRNA in situ hybridization. Panels A-C: grn1 is expressed in the intestine (arrow), swim bladder, and more abundantly in the head kidneys (arrowheads) than in the pronephric ducts. Panels D-F: grn2 is expressed similarly to grn1 in the head kidneys (arrowheads) and pronephric ducts, but is undetected in the intestine. In contrast, grn2 is strongly expressed in the brain and the branchial jaw region (compare A with D, and C with F), is distributed in a punctuate pattern along the ventral region of the animal in presumed myeloid progenitors (asterisks in D), and often found in randomly dispersed leukocytes (large cells in F). Panel G: Hybrid grn is found exclusively in the proctodeum. Panel H: The sense riboprobe to ASgrn1-2 (devoid of the tzf sequence) recapitulates the combined expression patterns for grn1 and grn2. Panels (I-N): Sense riboprobes to grn1 (I,J) or grn2 (K,L), but not hybrid grn (M), show that antisense transcription occurs in the jaw region (arrows in I and K), the swim bladder, and in the mid-region of the intestine, in a pattern identical to that observed for the antisense probe corresponding to ASgrn1-2 (N). B, E: dorsal views; C, F, I, K: ventral views. For each target mRNA, the use of corresponding sense and antisense (AS) riboprobes is indicated. OC, presumed ossification center; HK, head kidney; PD, pronephric duct; SB, swim bladder. EXPRESSION / LABELING:
|