- Title
-
Enhancing acute kidney injury regeneration by promoting cellular dedifferentiation in zebrafish
- Authors
- Skvarca, L.B., Han, H.I., Espiritu, E.B., Missinato, M.A., Rochon, E.R., McDaniels, M.D., Bais, A.S., Roman, B.L., Waxman, J.S., Watkins, S.C., Davidson, A.J., Tsang, M., Hukriede, N.A.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
|
|
|
|
|
|
|
|
|

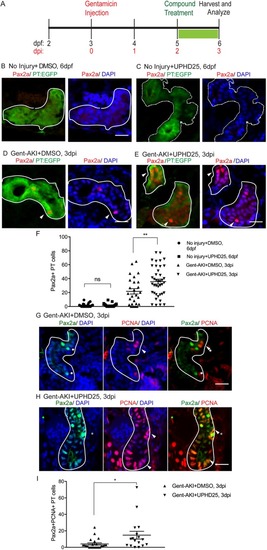
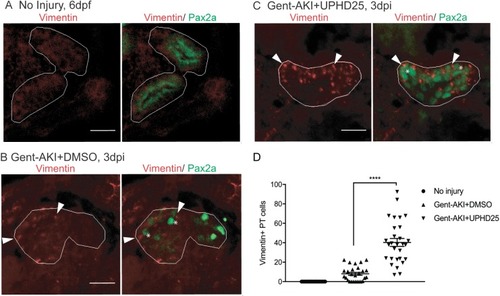
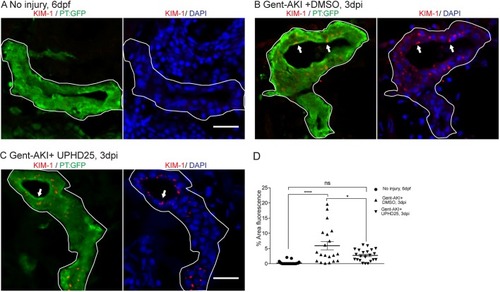
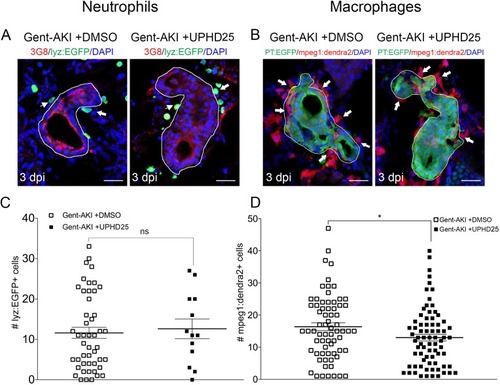
ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

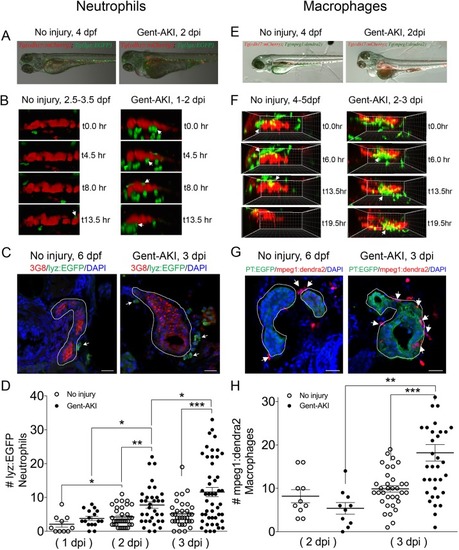
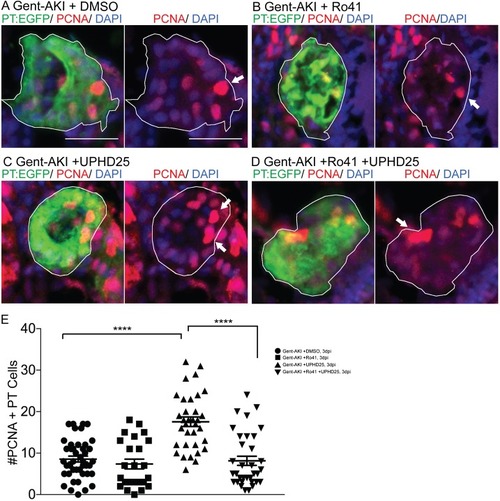
ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|