- Title
-
How Surrogate and Chemical Genetics in Model Organisms Can Suggest Therapies for Human Genetic Diseases
- Authors
- Strynatka, K.A., Gurrola-Gal, M.C., Berman, J.N., McMaster, C.R.
- Source
- Full text @ Genetics
|
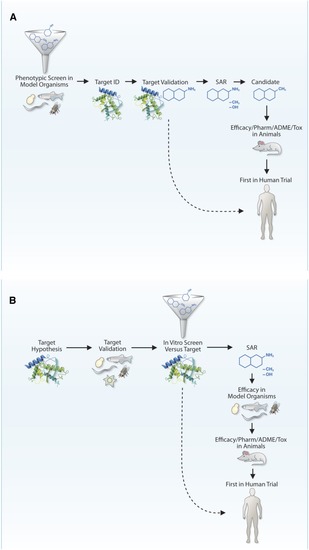
Outside-in and inside-out approaches to drug discovery for genetic diseases. (A) Outside-in screens start with a small-molecule screen |
|
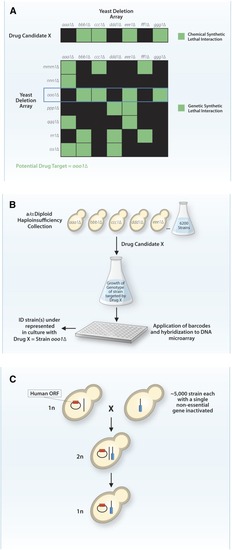
Application of yeast for drug discovery for genetic diseases. (A) A small molecule (drug) is screened |
|
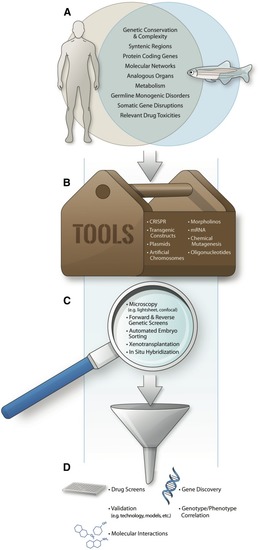
Applications of the zebrafish model in the study of inherited and acquired human genetic diseases. (A) Zebrafish and humans share a high degree of genetic conservation, including chromosome synteny and protein-coding genes. Molecular networks are also conserved, permitting use of zebrafish for investigating downstream pathways implicated in both inherited human diseases and somatic mutations like those found in cancer. Moreover, as a vertebrate model, the presence of analogous organs and conserved metabolism enable functional studies of the impact of genetic mutations on specific tissues and predictive readouts of therapeutic responses to drug administration. (B) The zebrafish is amenable to a number of genetic tools, including clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-based genome editing and transgenic technology, to introduce specific human mutations, including inserting oligonucleotides into the zebrafish genome. Other tools—including the facile introduction of plasmid DNA, BACs, and YACs; mRNA for transient gene overexpression studies and morpholinos for transient gene knockdown; and chemical mutagenesis—make the zebrafish a versatile model system for gene manipulation and phenotypic readouts. (C) Genetically modified zebrafish are easily surveyed using light and fluorescent microscopy, which can be enhanced with the availability of Lightsheet technology and confocal imaging. Whole-mount |