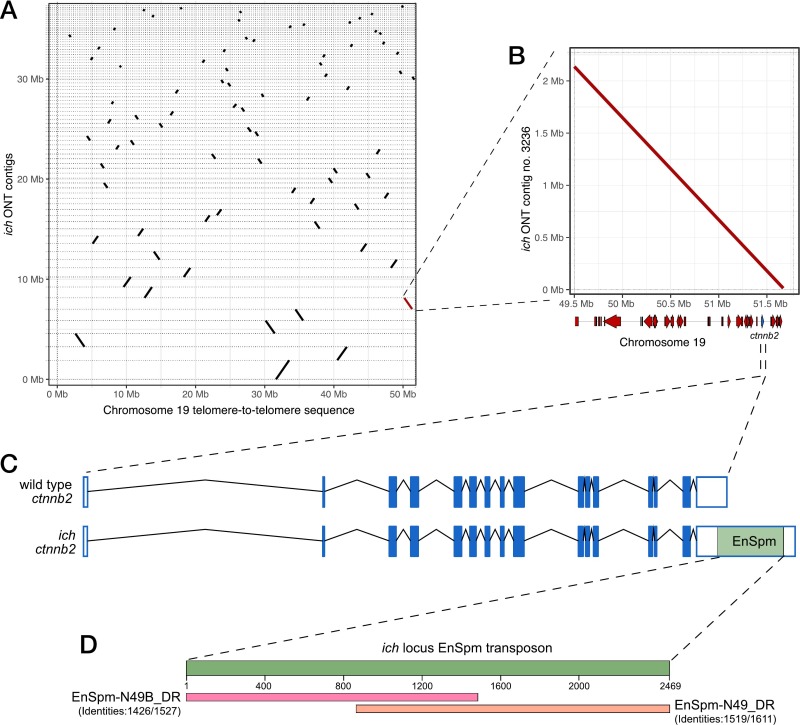
Fig. 1 Long-read sequencing identifies an EnSpm transposon in the 3’UTR of ctnnb2 in ich animals. (A) Dotplot representation of different ich contigs that could be aligned to the “telomere-to-telomere” sequence of the zebrafish chromosome 19. The highlighted (red) contig contains the genomic region of the ctnnb2 gene. (B) Dotplot comparison of the relevant ich contig and the telomeric portion of chromosome 19 reveals no genomic rearrangement in this region in ich animals. Gene position schematics are visible along the horizontal axis. (C) Comparison of wild-type and ich-specific ctnnb2 sequences showing the position of the EnSpm-type transposon insertion. (D) BLAST results of the ich-specific transposon in the FishTEDB [40].
Reprinted from Biochimica et biophysica acta. Gene regulatory mechanisms, , Varga, Z., Kagan, F., Maegawa, S., Nagy, Á., Okendo, J., Burgess, S.M., Weinberg, E.S., Varga, M., Transposon insertion causes ctnnb2 transcript instability that results in the maternal effect zebrafish ichabod (ich) mutation, 195104195104, Copyright (2025) with permission from Elsevier. Full text @ BBA Gene Regulatory Mechanisms