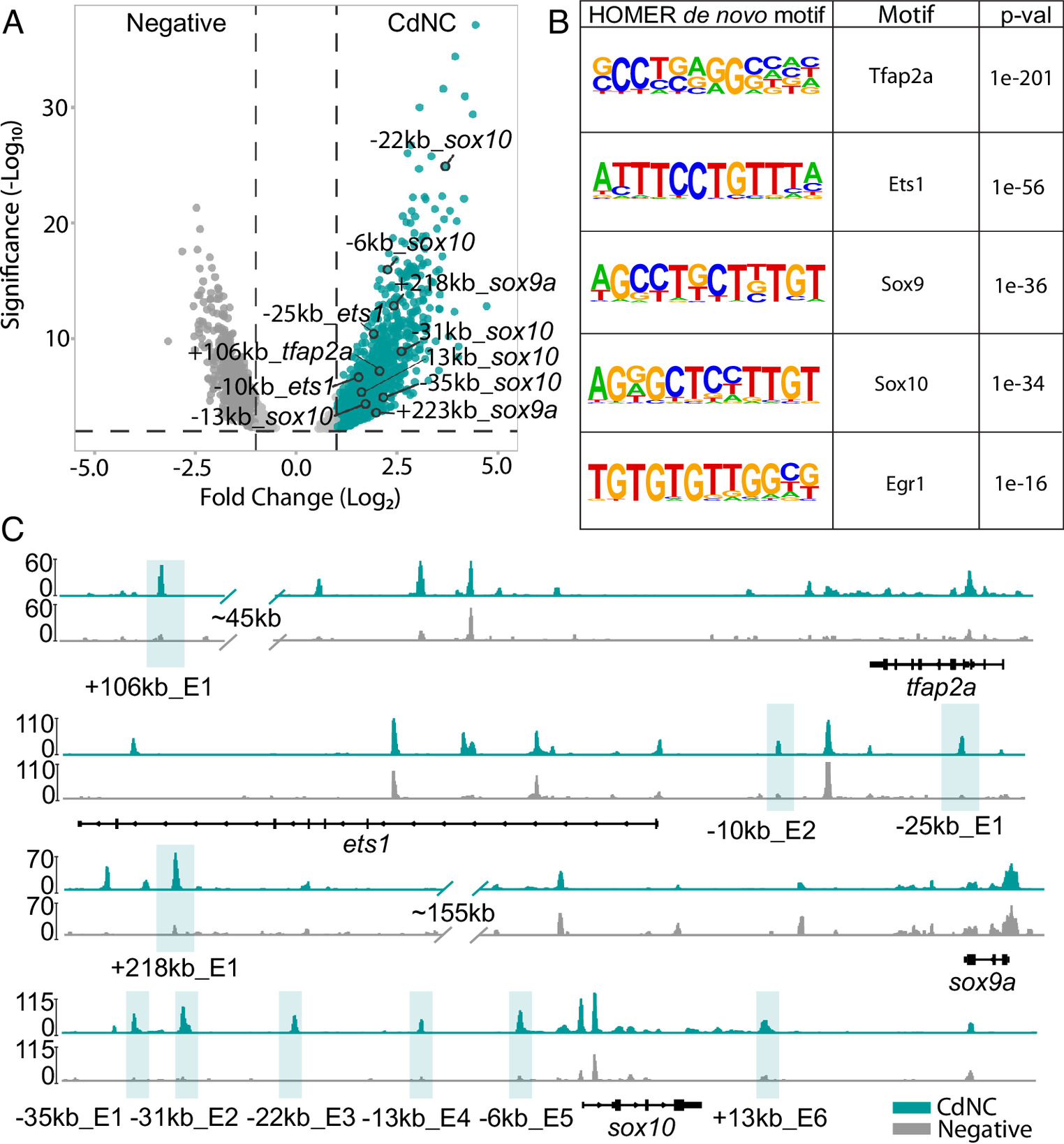
Fig. 2 ATAC-seq and differential chromatin accessibility and transcription factor motif analysis uncovers putative enhancers and transcriptional regulators unique to the CdNC. (A) Volcano plot of merged peaks of sorted bulkATAC-seq shows differentially accessible regions in CdNC (mCherry+) and non-NC/negative cells (mCherry-). (B) Results of HOMER de novo motif analysis showing motif, transcription factor, and associated P-value. (C) IGV genome browser tracks of the ets1, tfap2a, sox9a, and sox10 loci. Highlighted peaks represent putative enhancer regions called by Diffbind (turquoise track = CdNC, gray track = negative).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA