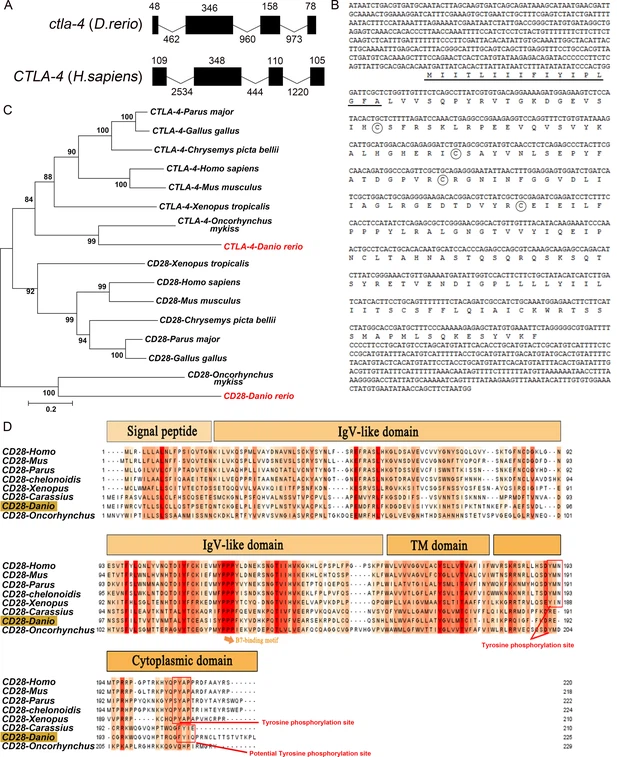
Fig. 1 - Supplemental 1 The organization, sequence, and phylogenetic analysis of zebrafish Cytotoxic T lymphocyte antigen-4 (ctla-4) and cd28 genes. (A) Comparison of the intron/exon organizations of ctla-4 gene in zebrafish and humans. Exons and introns are shown with black boxes and lines, and their sizes are indicated by the numbers found above and below the sequences respectively. (B) The nucleotide and amino acid sequences of ctla-4 gene and Ctla-4 protein. The underline indicates the signal peptide, the circles represent the conserved cysteine residues. (C) Phylogenetic analysis of the relationship of Ctla-4 and Cd28 between zebrafish and other species. An unrooted phylogenetic tree was constructed through the neighbor-joining method, based on amino acid sequence alignments generated by ClustalX. Bootstrap confidence values, derived from 500 replicates, are indicated as percentages at each node. (D) Alignment of the Cd28 homologs from different species generated with ClustalX and Jalview. The conserved and partially conserved amino acid residues in each species are colored in hues graded from orange to red, respectively. The conserved functional motifs, such as B7-binding motif, tyrosine phosphorylation site, and potential tyrosine phosphorylation site, were indicated separately. The signal peptide, IgV-like domain, transmembrane (TM) domain, and cytoplasmic domain were marked at the top of the sequence.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife