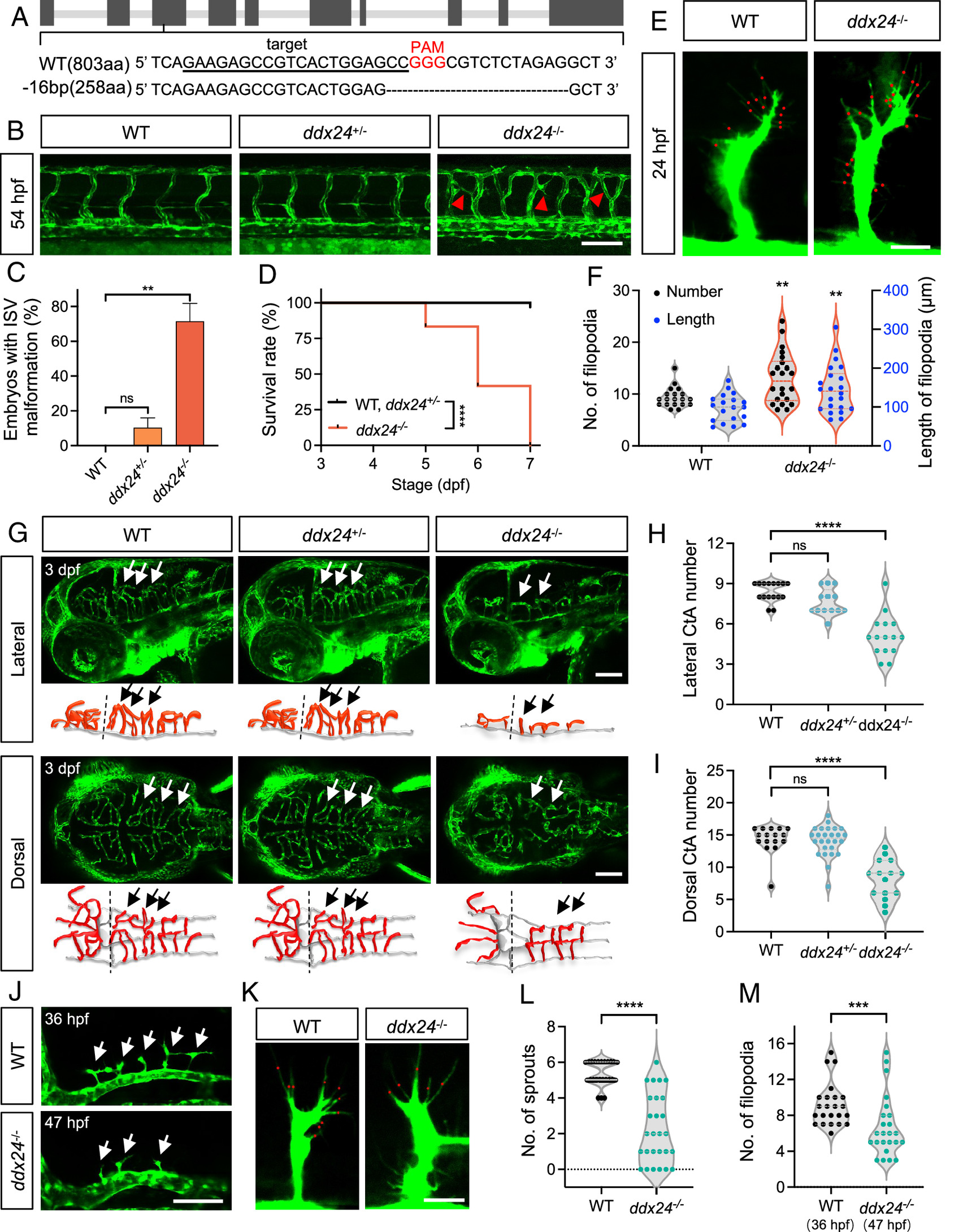
Fig. 2 Loss of Ddx24 promotes endothelial cell branching in the trunk and impairs sprouting in the brain of zebrafish. (A) CRISPR/Cas9 target site in ddx24 locus with 16-bp deletion (PAM in red). (B) Representative confocal images of trunk vasculature in WT, ddx24+/-, and ddx24−/− embryos at 54 hpf. Red arrowheads indicate ectopic ISV branches. (C) Quantification of the embryo with ectopic ISV branches. n = 29, 40, 23 embryos. (D) Kaplan–Meier survival curves of WT, ddx24+/-, and ddx24−/− zebrafish. n = 18, 36, 12 embryos. (E) Representative high-magnification confocal images of ISV sprouts in WT and ddx24−/− embryos at 24 hpf. The red dots point to the tip cell filopodia. (F) Quantification of filopodia number and length. n = 19 for WT, n = 22 for ddx24−/− embryos. (G) Representative confocal images and schematic diagram of hindbrain vasculature in WT, ddx24+/-, and ddx24−/− embryos at 3 dpf. Arrows indicate CtAs. (H and I), Quantification of CtAs. Lateral view number of CtAs (H, n = 16, 13, 15 embryos) and dorsal view number of CtAs (I, n = 16, 26, 15 embryos) were determined. (J) Representative confocal images of hindbrain vasculature in WT (36 hpf) and ddx24−/− (47 hpf) embryos. Arrows indicate CtAs. (K) Representative high-magnification confocal images of CtA sprouts in WT (36 hpf) and ddx24−/− (47 hpf) embryos. The red dots point to the tip cell filopodia. (L and M), Quantification of CtA sprouts (L, n = 24, 27 embryos) and filopodia per CtA (M, n = 23, 24 embryos). [Scale bars, 100 μm in (B, G, and J); 20 μm in (E and K).] Data are represented as mean ± SD. ns, not significant, **P < 0.01, ***P < 0.001 and ****P < 0.0001, as assessed by one-way ANOVA (C), log-rank (Mantel–Cox) test (D) parametric two-tailed Student’s t test (F), nonparametric Kruskal–Wallis test (H and I) and nonparametric Mann–Whitney test (L and M).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA