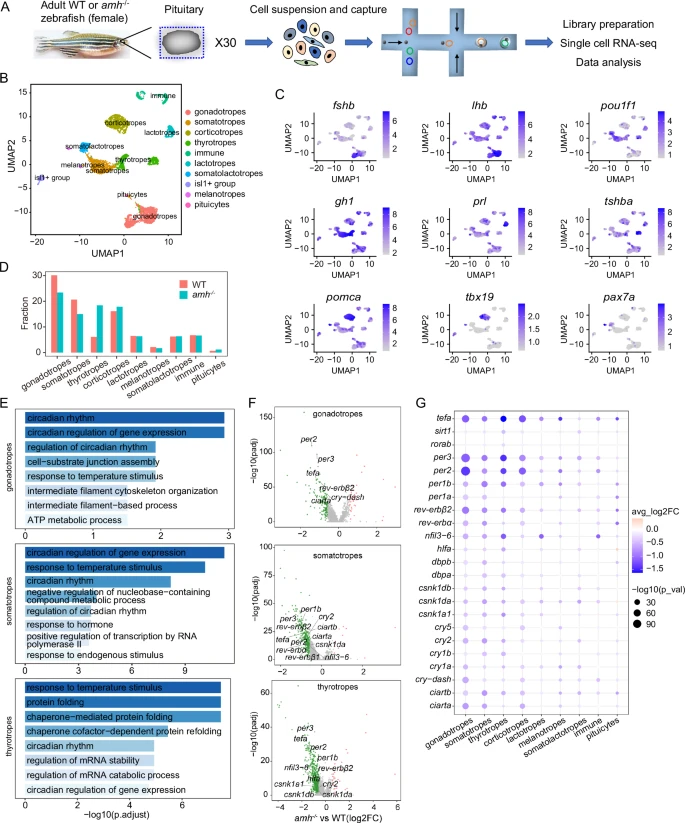
Fig. 3 Cell type-specific regulation of the molecular clock by Amh was revealed by single-cell transcriptome analysis of the pituitary.A Schematic overview of the process of cell isolation and single-cell RNA-seq analysis of the zebrafish pituitary using the 10X Genomics platform (30 pituitaries in each group, WT and amh−/−). B UMAP visualization shows the unsupervised clustering of the aggregated results of two scRNA-seq experiments, revealing 10 major clusters of pituitary cells present in zebrafish. Each dot represents one cell, and colours represent cell clusters as indicated. C UMAP visualization of cluster marker genes. UMAP, uniform manifold approximation and projection. D Bar graph showing the percentages of each cell cluster in the pituitaries of WT and amh−/− zebrafish. E Gene ontology enrichment for the downregulated genes from the major affected pituitary cell populations of amh−/− zebrafish. The top 8 terms for biological process (BP) were shown. Hypergeometric test and Benjamini-Hochberg method used for multiple comparisons correction. F Volcano plot showing differential circadian clock gene expression between amh−/− and WT zebrafish in the major affected pituitary cell lineages. G Dot plot showing the expression changes in circadian clock genes across the pituitary cell clusters from WT and amh−/− female zebrafish. For F, G two-sided unpaired Wilcoxon test and adjustments were made for multiple comparisons. Source data are provided as a Source Data file.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Commun.