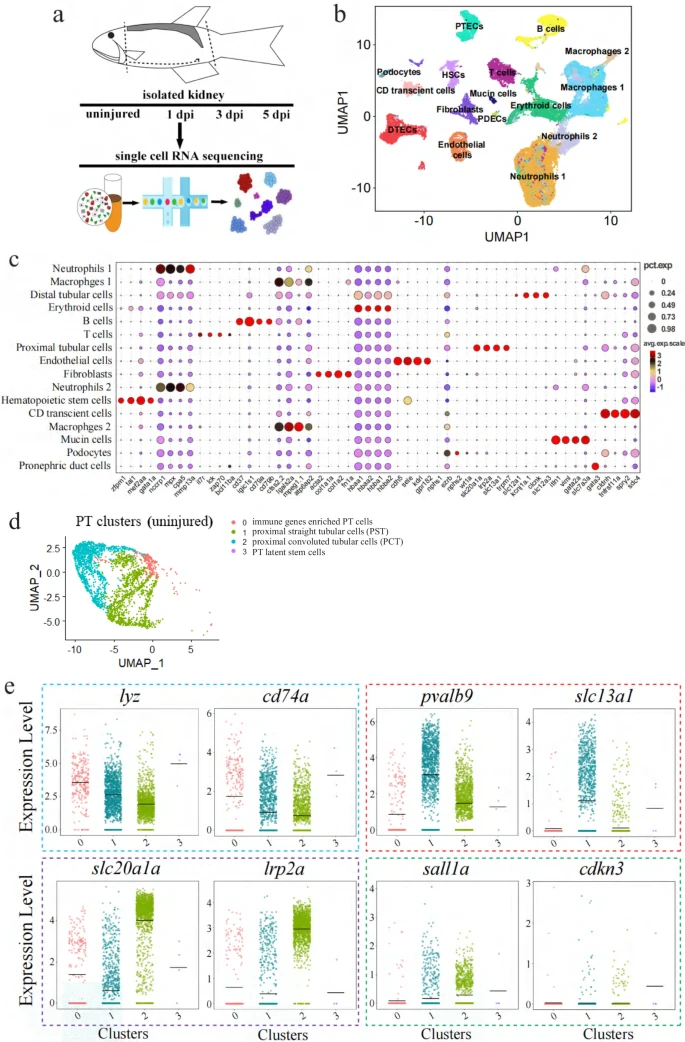
Fig. 1 scRNA-seq data analysis of zebrafish kidneys.a Schematics of the study design. b UMAP embedding of scRNA-seq data. Using kidney marker genes, cells were annotated into the indicated 16 major clusters. PTECs: proximal tubular epithelial cells; DTECs: distal tubular epithelial cells; B cells: B lymphocytes; T cells: T lymphocytes; PDECs: pronephric duct cells; PD; podocytes; HSCs: hematopoietic stem cells. c Bubble plot displaying the cluster-enriched marker genes. d UMAP projection of uninjured/wild-type proximal tubular cells (PT), demonstrating four sub-clusters. Subcluster 0: the immune gene-enriched cells; subcluster 1: proximal straight tubular cells (PST); subcluster 2: proximal convoluted tubular cells (PCT); sub-cluster 3: the resident stem cell population. e Violin plots grouped by meta-clusters, demonstrating the subcluster-specific expression of the indicated genes. The dashed colored boxes were used to indicate the markers of the four subclusters. 0–3 of the X-axis: the subclusters 0, 1, 2, 3.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Commun Biol