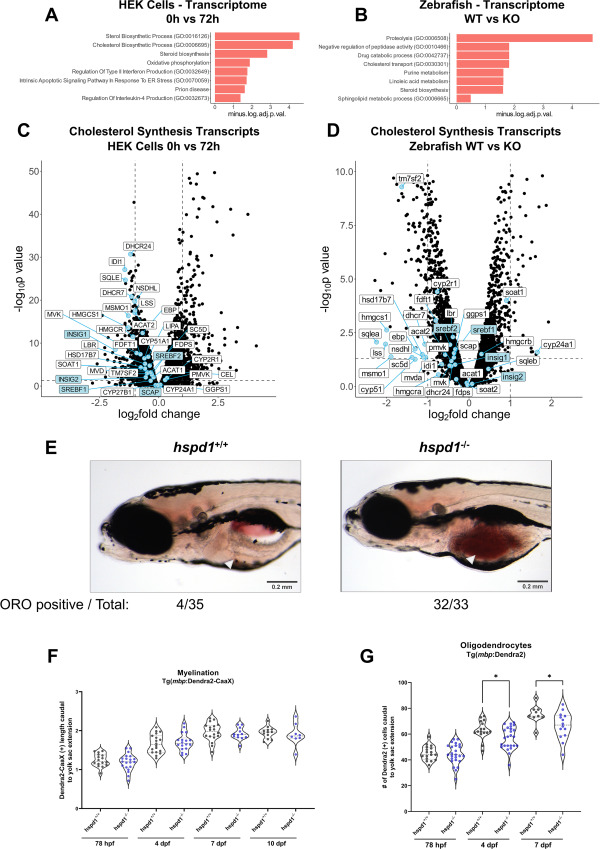
Fig. 6 Zebrafish Hsp60 knockout larvae have altered lipid metabolism and oligodendrocyte counts (A–B) Enrichment analysis of differentially expressed genes of D423A cells at 0 h in comparison to 72 h (A) and in 5 dpf hspd1−/− larvae in comparison to hspd1+/+ larvae (B). Performed on EnrichR at gene name level for proteomics and transcriptomics, GO biological process and KEGG terms are included. The enriched terms are shown as a negative log10 of adjusted p-values. Adjusted p-values <0.05 were considered significant. (C–D) Volcano plots showing the transcriptomic regulation of genes involved in cholesterol synthesis pathways in D423A cells induced for 72 h (C) and in 5 dpf hspd1−/− larvae (D). Cholesterol synthesis genes and regulators are displayed as blue points; the labels for the regulating system genes are shaded in blue. (E) Whole-mount Oil Red O staining of 8 dpf zebrafish larvae shows lipid accumulation and altered lipid composition in the yolk of hspd1−/− larvae compared to hspd1+/+ larvae. (F) The length of Dendra2-CaaX(+) signal was measured caudal to yolk sac extension at 78 hpf, 4 dpf, 7 dpf, and 10 dpf. Larvae were generated from hspd1+/− to Tg(mbp:Dendra2-CaaX) mating. (G) The number of mature oligodendrocytes was quantified as caudal to yolk sac extension Dendra2-positive cells at 78 hpf, 4 dpf, and 7 dpf. Larvae were generated from hspd1+/− to Tg(mbp:Dendra2) mating. Abbreviations: TCA, tricarboxylic acid; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. See also Figure 1 , Figure 2 , Figure 4 .
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Mol Metab