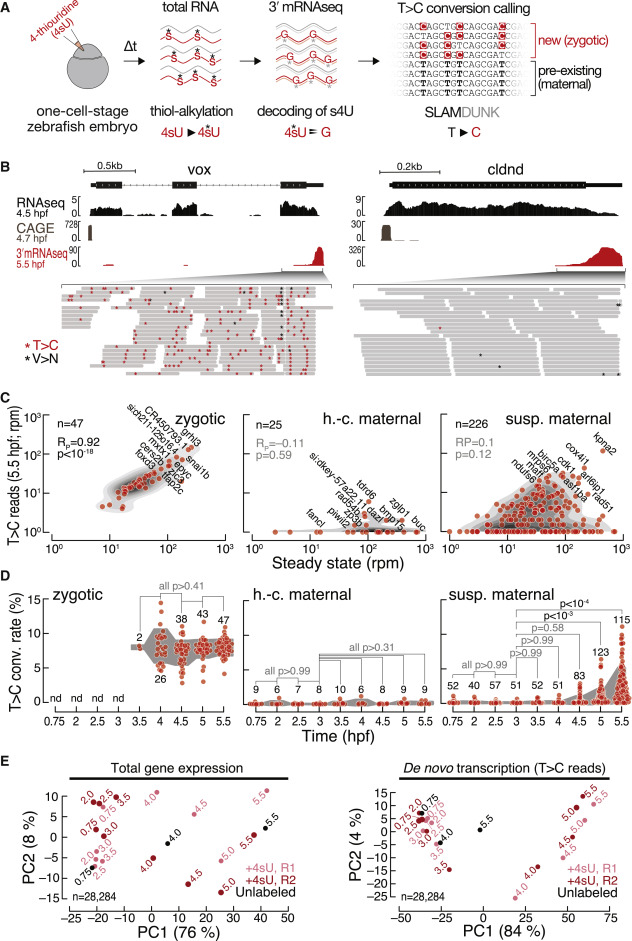
Fig. 1 3′ mRNA SLAMseq in zebrafish embryos distinguishes zygotic from maternally deposited mRNA (A) Zebrafish embryos were injected with 4-thiouridine (4sU; 1.5 mM) at the 1-cell stage and development proceeded for a defined time (Δt), followed by total RNA preparation and SLAMseq employing 3′ mRNA sequencing. Zygotic transcripts were distinguished from maternal mRNAs by the presence of T>C conversions at sites of 4sU incorporation as identified by SLAMDUNK.21 (B) Representative genome browser screenshot of the zygotic gene ventral homeobox (vox; left) and the maternal gene claudin d (cldnd; right). Tracks for poly(A)-selected RNA-seq (top), cap analysis gene expression (CAGE; middle; remapped from Haberle et al.14), and 3′ mRNA SLAMseq (bottom), sampled at the indicated developmental stage (hours post-fertilization [hpf]), are shown. Zoom in shows individual reads of 3′ mRNA SLAMseq datasets mapping to the respective 3′ UTR counting windows. Asterisks indicate T>C conversions (T>C; red) or any other conversions (V>N; black). (C) Comparison of de novo transcription (T>C reads in reads per million [rpm]) and gene expression (in rpm) for pre-defined zygotic (n = 47), high-confidence (h.-c.; n = 25), and suspected (susp.; n = 226) maternal genes at 5.5 hpf. Pearson correlation coefficient (RP) and associated p values (p) are shown. (D) T>C conversion (conv.) rate for transcripts defined in (C) during MZT (time in hpf). Individual genes for which a confident average conversion rate could be derived from two independent biological replicates (red, n = number of genes) and interquartile range (gray area) are shown. p values (Kruskal-Wallis and Dunn’s multiple comparison test) are indicated; nd, not detected. (E) Principal-component analyses of unfiltered control (unlabeled; black) and two independent 3′ mRNA SLAMseq (+4sU; R1 light and R2 dark red) experiments at the indicated developmental time points (in hpf). Gene expression (left); de novo transcription (T>C reads) (right). The variance of each principal component is indicated in a percentage. n, number of genes considered for the analysis.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.