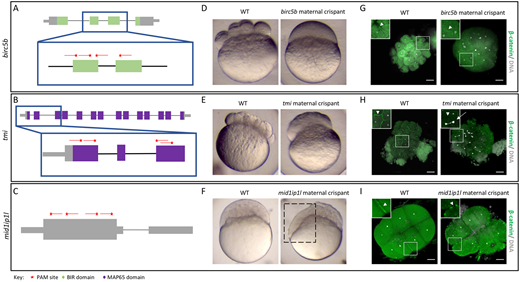
Fig. 2 Identification of known maternal-effect genes using maternal crispants. (A-I) Known maternal-effect phenotypes resulting from the mutagenesis of three genes, birc5b (A,D,G), tmi (B,E,H) and mid1ip1l (C,F,I), were replicated using the maternal crispant technique. (A-C) Gene structure diagrams of birc5b (A), tmi (B) and mid1ip1l (C) showing the target sites of guide RNAs (red lines) and PAM sites (red stars). (D-F) Representative comparisons of a live wild-type (WT) embryo with live time-matched maternal crispant embryos of birc5b [D; 75 min post-fertilization (mpf)], tmi (E; 75 mpf) and midip1il (F; 150 mpf), showing defects in cytokinesis identical to previous studies. The mid1ip1l maternal crispant embryo shown exhibits a partially syncytial phenotype (box in F). (G-I) Immunohistochemistry labeling of β-catenin and DAPI staining in birc5b (G; 90 mpf), tmi (H; 90 mpf) and mid1ip1l (I; 75 mpf) maternal crispants showing the lack of accumulation of β-catenin in mature furrows, confirming a failure in furrow formation in spite of DNA replication. mid1ip1l maternal crispants contain ectopic β-catenin-containing vesicles near the cortex. In all immunohistochemistry labeling, insets show high magnification images of the boxed areas, highlighting the differences in β-catenin localization (arrowheads) and asymmetric DNA segregation (arrow). Note that there is some embryo size variation caused by methanol storage of embryos. Scale bars: 100 μm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development