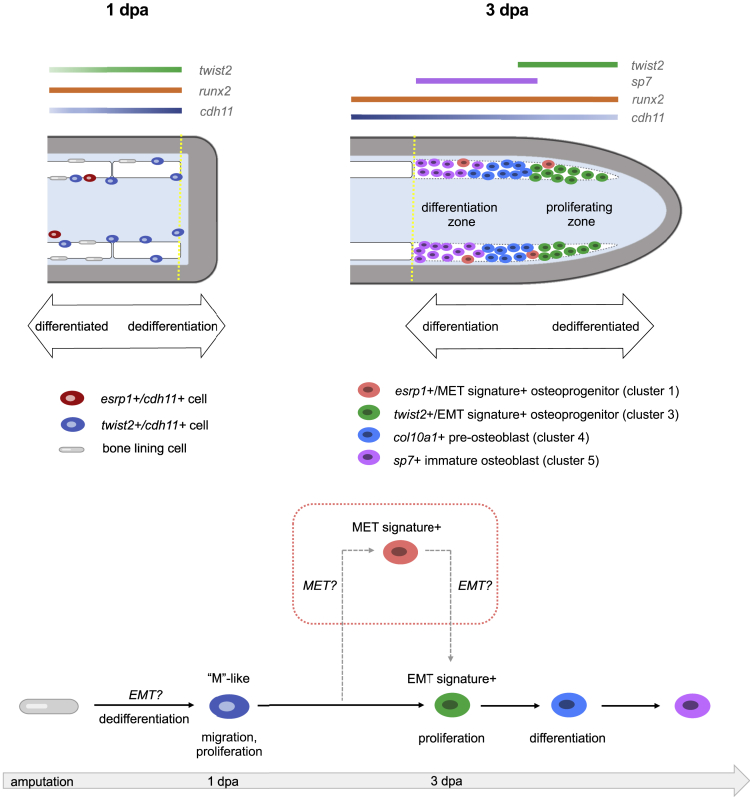
Fig. 6
Schematics showing predicted spatial locations of the different osteoprogenitors from osteoblast2 within the fin (top) and predicted model of osteoblastic de- and redifferentiation during fin regeneration (bottom)
Amputation plane is indicated with a dashed yellow line. (top left) At 1 dpa, dedifferentiated osteoblasts expressing cdh11 and twist2 are observed along the native bone, at fin ray joints, and at the amputation stump. Elongated bone lining cells are still present but are twist2-/cdh11-. esrp1+/cdh11+ cells are occasionally found near the native bone. (top right) At 3 dpa, osteoprogenitors in the blastema are located along the lateral edges of the mesenchymal compartment as indicated by the dashed white region. Early cycling osteoprogenitors expressing a strong EMT gene signature are distally located. These cells also exhibit robust expression of runx2a and twist2, and comparably less cdh11 than in proximal regions of the regenerate. Proximal osteoprogenitors exhibit strong cdh11 expression and have begun maturing into differentiated osteoblasts, first expressing col10a1, then sp7. Non-cycling osteoprogenitors, some of which express a strong MET gene signature, are sporadically located among both the proximal and distal osteoprogenitors. (bottom) Model of osteoblastic de- and redifferentiation during fin regeneration. Bone lining cells dedifferentiate into twist2+/cdh11+ cells, which are mesenchymal-like (“M”-like). These cells then migrate into the blastema and proliferate. A majority of the cells then follow the solid black arrows and progress through redifferentiation and become mature osteoblasts to regenerate bone. Some of the cells follow the dashed gray arrows, first expressing MET components before expressing EMT components, prior to differentiating into mature osteoblasts.