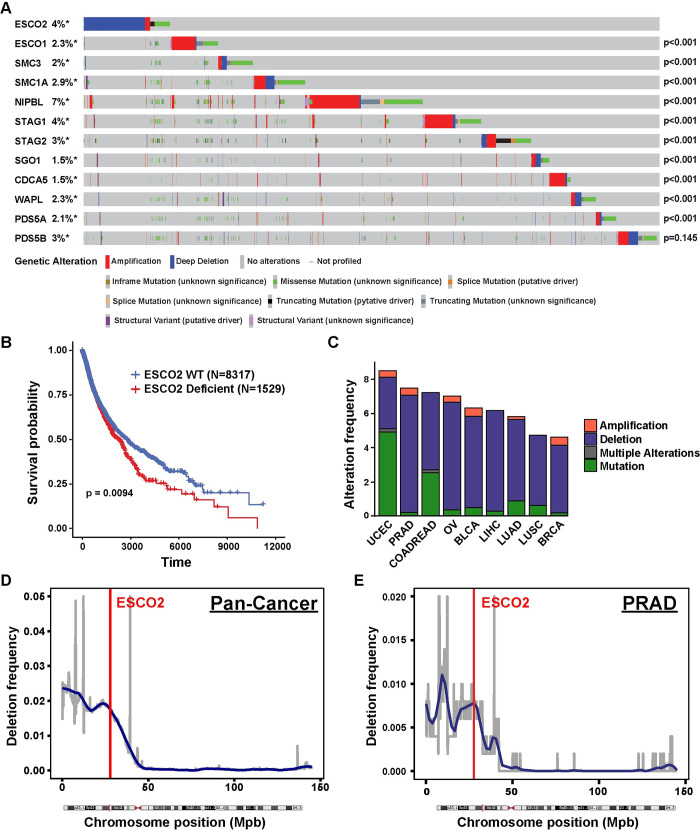
Fig 1
(A) Oncoprint plot of genetic alterations in ESCO2 and other SCC-associated genes in TCGA dataset. N = 10950. Number of samples with each mutation: N(ESCO2) = 404, N(ESCO1) = 252, N(SMC3) = 216, N(SMC1A) = 322, N(NIPBL) = 743, N(STAG1) = 429, N(STAG2) = 374, N(SGO1) = 165, N(CDCA5) = 159, N(WAPL) = 248, N(PDS5A) = 227 and N(PDS5B) = 375. P-values shown in the figure indicate significance of co-occurrence with ESCO2 from one-sided Fisher Exact test. (B) Kaplan-Meier survival analysis of patients with ESCO2 deficient (N = 1529) versus ESCO2 WT (N = 8317) tumors within TCGA dataset samples. P-value was determined by log-rank test. (C) Stacked bar plot indicating the percentage of patients with an ESCO2 deletion (blue), amplification (red) or mutation (green) and multiple alterations (grey) in uterine corpus endometrioid (UCEC), prostate (PRAD), colorectal (COADREAD), ovarian (OV), bladder (BLCA), liver (LIHC), lung adenocarcinoma (LUAD), lung squamous (LUSC), and breast cancer (BRCA) in the TCGA dataset. Cancer type is ordered based on the genetic alteration frequency. Number of samples with mutation/ total samples in each cancer type: 45/529 UCEC, 37/494 PRAD, 43/594 COADREAD, 41/584 OV, 26/411 BLCA, 23/372 LIHC, 33/566 LUAD, 23/487 LUSC, 50/1084 BRCA. The frequency of deletion in 100 kbp windows throughout Chromosome 8 in 1,111 pan-cancer type (Figure D, N = 11203) and prostate adenocarcinoma (PRAD, Figure E, N = 502) cancer patients. A region is considered as deleted if Log2 (Copy Number/2) < -1. Blue line shows smoothed deletion frequency. Red vertical line indicates the locus of ESCO2 gene.