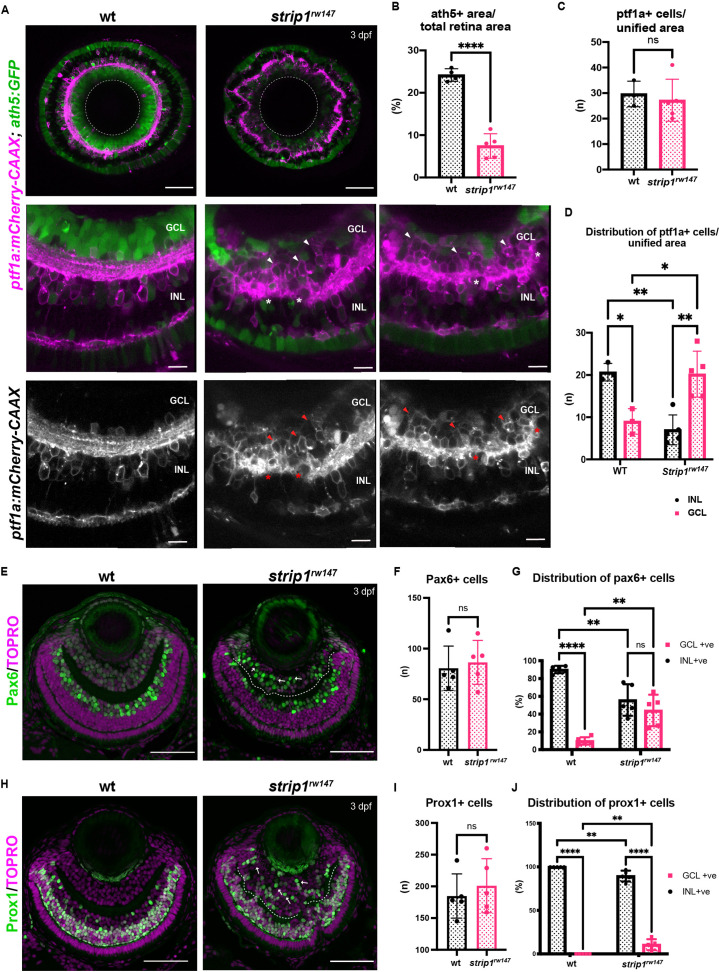
Figure 2
(A) Confocal sections of wild-type and strip1rw147 mutant retinas combined with the transgenic line Tg[ath5:GFP; ptf1a:mCherry-CAAX] to label RGCs and amacrine cells (ACs). Middle panels represent higher magnification. Lower panels show the magenta channel. Arrowheads indicate abnormal positioning of ptf1a+ ACs in the GCL. Asterisks show AC dendritic patterning defects. INL, inner nuclear layer; GCL, retinal ganglion cell layer. Scale bars, 50 μm (upper panels) and 10 μm (middle and lower panels). (B) Percentage of ath5+ area relative to total retinal area. Student’s t-test with Welch’s correction, n ≥ 4. (C) AC numbers per unified retinal area (8500 μm2). Student’s t-test with Welch’s correction, n ≥ 3. (D) Distribution of ACs (GCL or INL) per unified retinal area (8500 μm2). Two-way analysis of variance (ANOVA) with the Tukey multiple comparison test, n ≥ 3. (E) Wild-type and strip1rw147 mutant retinas at 3 dpf labeled with anti-Pax6 antibody which strongly labels ACs. Arrows indicate strong Pax6+ cells that infiltrate the GCL. Nuclei are stained with TOPRO3. Scale bar, 50 μm. (F) The number of strong Pax6+ cells per retina. Student’s t-test with Welch’s correction, n = 5. (G) Percentage of strong Pax6+ cells (GCL+ or INL+) to the total number of strong Pax6+ cells. Two-way ANOVA with the Tukey multiple comparison test, n = 5. (H) Wild-type and strip1rw147 mutant retinas at 3 dpf labeled with anti-Prox1 antibody. Arrows indicate Prox1+ cells that infiltrate the GCL. Nuclei are stained with TOPRO3. Scale bar, 50 μm. (I) The number of Prox1+ cells per retina. Student’s t-test with Welch’s correction, n = 5. (J) Percentage of Prox1+ cells (GCL+ or INL+) to the total number of Prox1+ cells. Two-way ANOVA with the Tukey multiple comparison test, n = 5. For all graphs, data are represented as means ± standard deviation (SD). ns, not significant, *p < 0.05, **p < 0.01, and ****p < 0.0001.
Retinal ganglion cells (RGCs) are reduced and INL cells infiltrate the GCL in strip1 mutants.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife