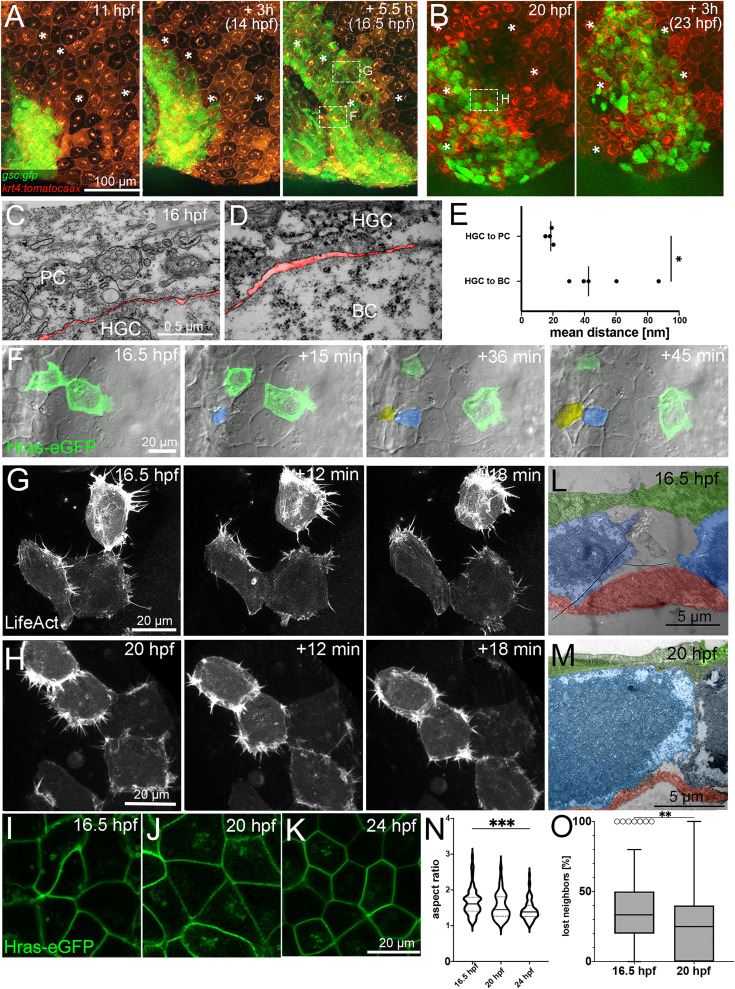
Fig. 5 A,B. Fluorescent stills from a time-lapse recording (Supplementary movies 2 and 3) of an embryo transgenic for Tg(-1.8gsc:gfp) and Tg(krt4:tdtomatocaax), starting at 11 hpf (A) and 20 hpf (B). ∗ indicate identical peridermal cells at each time point. Boxed regions in images at 16.5 hpf and 20 hpf indicate regions for which subsequent time-lapse recordings are presented in (F,G,H) at higher magnifications. C,D. TEM sections of contact zones between hatching gland cell (HGC) and peridermal cell (PC) (C) and between HGC and basal keratinocyte (BC) (D) at 16 hpf. The interspace between cells is false-colored in red. E. Quantification of the spaces between hatching gland cells and peridermal and basal cells, respectively. Each dot represents one cell-cell contact. N = 2 embryos, n = 4–5 cells. ∗ indicates p value of 0.0213 determined via a Student’s t-test. F. Bright-field and fluorescent overlay stills from a time-lapse recording (Supplementary movie 4) of multi-layered zone as indicated in third image of (A), in this case of a wild-type embryo with two transplanted GFP-positive cells transgenic for Tg(Ola.Actb:Hsa.hras-egfp), starting at 16.5 hpf. Cells intercalating from below are false-colored in yellow and blue. G,H. Fluorescent stills from time-lapse recordings (Supplementary movies 5 and 6) of HGCs in mono-layered zone as indicated in third image of A (G) and first image of B (H) of wild-type embryos injected with a 1.8gsc:LifeAct-Ruby plasmid labeling active actin cytoskeletal rearrangements, starting at 16.5 hpf (G) or 20 hpf (H). L,M. TEM sections through HGCs (false-colored in blue) between peridermal cells (PC, false-colored in green) and basal keratinocytes (BC, false-colored in red) at 16.5 hpf (L) or 20 hpf (M). I–K. Confocal images of cell membranes of HGCs of embryos transgenic for Tg(Ola.Actb:Hsa.hras-egfp) to label cell membranes at 16.5 hpf (I), 20 hpf (J), or 24 hpf (K). N. Quantification of aspect ratio of HGCs shown in I–K in violin blot; N = 5–9 embryos, n = 39–72 cells; ∗∗∗ indicates p value of 0.0001 determined via one-way ANOVA and Tukey’s post hoc test, non-significant differences are not indicated. O. Tukey box and whiskers plot displaying the percentages of neighbors of HGCs that were lost during a tracking period of 50 min in embryos transgenic for Tg(-1.8gsc:gfp), starting at 16.5 hpf or 20 hpf (see also Fig. 6A and I). N = 3 embryos each, n = 28–57. ∗∗ indicates p value of 0.0013 determined via Student’s t-test.
Reprinted from Developmental Biology, 476, Hatzold, J., Wessendorf, H., Pogoda, H.M., Bloch, W., Hammerschmidt, M., The Kunitz-type serine protease inhibitor Spint2 is required for cellular cohesion, coordinated cell migration and cell survival during zebrafish hatching gland development, 148-170, Copyright (2021) with permission from Elsevier. Full text @ Dev. Biol.