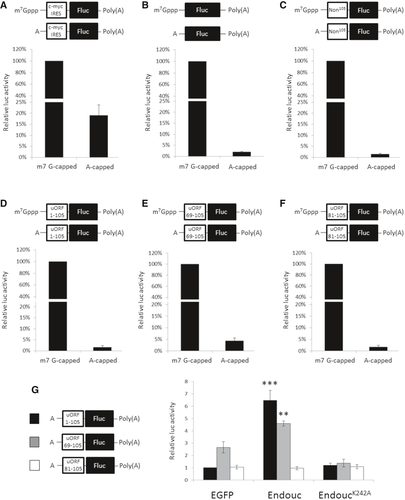
Fig. 10 A–F. Schematic representation of capped (m7G-capped) and A-capped (non-functional capped) monocistronic mRNAs with poly(A)-tail-encoded firefly luciferase (Fluc) and fused upstream with (A) c-myc IRES, (B) no insertion, (C) non-specific 105-nt luc RNA (Non105), (D) huORFchop-1-105-nt, (E) huORFchop-69-105-nt, and (F) huORFchop-81-105-nt as shown above each bar graph. The RNA transfection assay of each transcript equipped with m7G-capped or A-capped monocistronic mRNA in HEK293T cells was compared. To normalize the luc activities of capped and A-capped Fluc transcripts, the m7G-capped Renilla luc (Rluc) mRNA was used as an internal control. The luc activity of cells transfected with cap-monocistronic mRNA in each graph was set as 100%. G. The luc activity driven by the indicated A-capped monocistronic mRNA in the presence of Endouc or EndoucK242A in HEK293T cells. The m7G-capped Rluc mRNA was used as an internal control to normalize the activities of A-cap Fluc transcripts. The egfp mRNA was used as a negative control, and the luc activity of cells transfected with egfp mRNA and A-cap Fluc mRNA containing full-length huORFchop-1-105-nt was set to 1. The data were averaged from three independent experiments and presented as mean ± SD (n = 3). Student's t-test was used to determine significant differences between each group (**P < 0.005, ***P < 0.001).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ EMBO J.