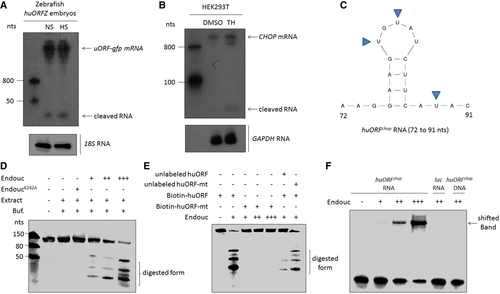
Fig. 7 Northern blot analysis to detect the endogenous huORFchop-tagged gfp (uORF-gfp) mRNA and its cleaved form in zebrafish embryos of transgenic line huORFZ treated with non-stress condition (NS) or heat-shocked (HS) stress. Zebrafish 18S rRNA served as an internal control. The RNA fragments cleaved from huORFchop transcript were detected. B. Northern blot analysis to detect endogenous CHOP mRNA in HEK293T cells treated with either DMSO (control group) or thapsigargin (TH). Human GAPDH RNA served as internal control. The RNA fragments cleaved from CHOP mRNA were detected. C. The predicted RNA sequences of Endouc recognition site on huORFchop transcript from 72 to 91 nts are illustrated. The predicted cleavage sites at uridylates (U) are indicated by arrowheads. D, E. The cleavage activity of Endouc was studied using materials presented at each lane. EndoucK242A: Endouc mutant; Extract: incubated in cell extracts; and Buf: reaction buffer. RNA probe without adding reaction buffer served as a negative control. The predicted cleavage sites (U) were mutated to generate mutant huORFchop transcript (unlabeled huORF-mt) and labeled by biotin (biotin-huORF-mt). Unlabeled competitor RNA was added in reactions at five times higher than that of the biotinylated probe. F. Electrophoretic mobility shifted assay of RNA. Biotin-labeled huORFchop transcript was reacted with the increased amount of Endouc. The positions of free RNA (free probe) and RNA–protein complex (shifted band) were indicated on the right. The luc 105-nt RNA and single-stranded huORFchop 105-nt DNA served as negative controls.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ EMBO J.