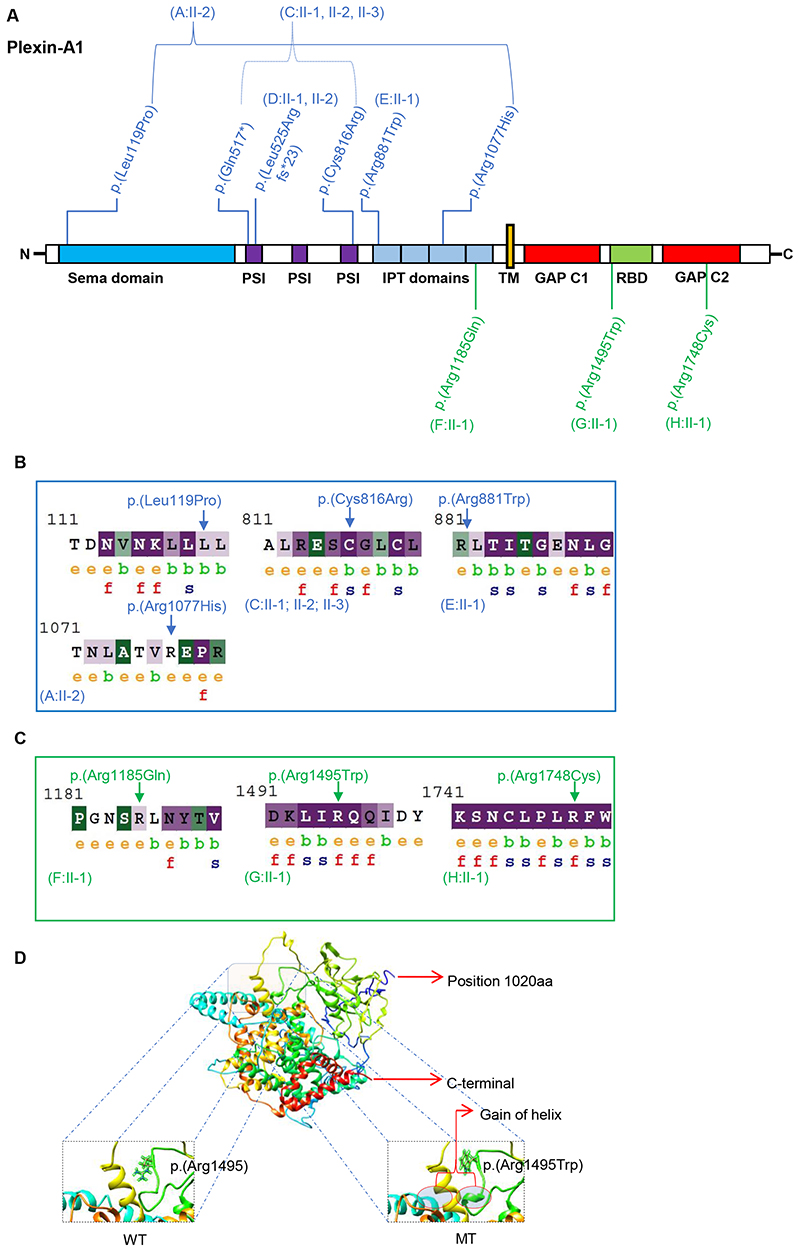
Figure 3 Fig. 3. Plexin-A1 variant distribution, conservation, and modeling. a Schematic protein domain structure adapted from St. Clair et al.5 and localization of Plexin-A1 variants. Note the nonrandom concentration of the biallelic variants (blue) in the extracellular domains and the monoallelic variants (green) toward the intracellular domains. GAP GTPase-activating protein, IPT Ig domain shared by plexins and transcription factors, PSI plexin-semaphorin-integrin domain, RBD Rho GTPase-binding domain, TM transmembrane region. b, c ConSurf analysis of the biallelic (b) and monoallelic (c) missense variants. The monoallelic variants (p.[Arg1185], p.[Arg1495], and p.[Arg1748]) are exposed suggesting functional residues in the protein. The intracellular residues p.(Arg1495) and p.(Arg1748) are evolutionarily conserved based on their ConSurf analysis suggesting that these residues react highly sensitively if altered. The biallelic missense variants (p.[Leu119Pro], p.[Cys816Arg], p.[Arg881Trp], p.[Arg1077His]) are noticed to be less conserved compared to the monoallelic variants with the p.(Cys816Arg) variant representing an exemption of this observation. e exposed, b buried, f functional, s structural. d 3D protein structure prediction shows replacement of the arginine long side chain by a flat aromatic ring of tryptophan (p.[Arg1495Trp]). Superimposition of the p.(Arg1495Trp) variant onto the wild-type structure showed that there is a gain of helix in the altered protein in close proximity to the site of variant. MT mutated, WT wild type.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Genet. Med.