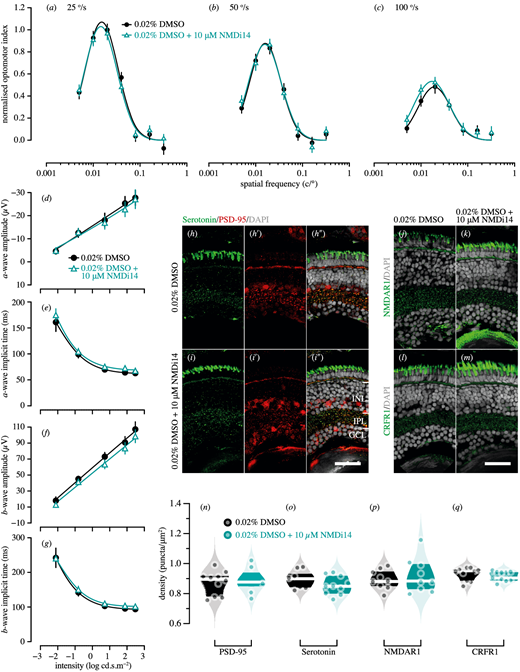
Fig. 8 Genetic compensation tests for pdzk1-KO mutants at 7 dpf. Spatial frequency tuning functions were fitted by a log-Gaussian model for control (pdzk1-KO mutants treated with 0.02% DMSO; black circles and lines) and experimental groups (pdzk1-KO mutants treated with 0.02% DMSO and 10-µM NMDi14; blue triangles and lines) at (a) 25°/s, (b) 50°/s, and (c) 100°/s. ERG results were shown as the group average (±SEM) of (d) a-wave amplitude, (e) a-wave implicit time, (f) b-wave amplitude, and (g) b-wave implicit time for pdzk1-KO mutants treated with 0.02% DMSO (filled black circles) and mutants treated with 0.02% DMSO and 10-µM NMDi14 (open cyan triangles). Data were compared using two-way ANOVA; there were no significant differences. Lines were fit by a four-parameter sigmoidal function. For histological assessment, (h–i″) serotonin and PSD-95 were co-labeled in green and red, respectively, for both groups; (j, k) NMDAR1and (l, m) CRFR1 were labeled in green. For all images, nuclei were stained with DAPI, shown in gray. Scale bars: 25 µm. Violin plots show densities in the IPL of (n) PSD-95, (o) serotonin, (p) NMDAR1, and (q) CRFR1 for mutants treated with 0.02% DMSO (black circles) and mutants treated with 0.02% DMSO and 10-µM NMDi14 (cyan circles). Points represent data from individual retinas. White lines represent medians, and dark bands indicate interquartile ranges. Unpaired t-tests were performed with 10 or 11 retinas per group (see Supplementary Table S6).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Invest. Ophthalmol. Vis. Sci.