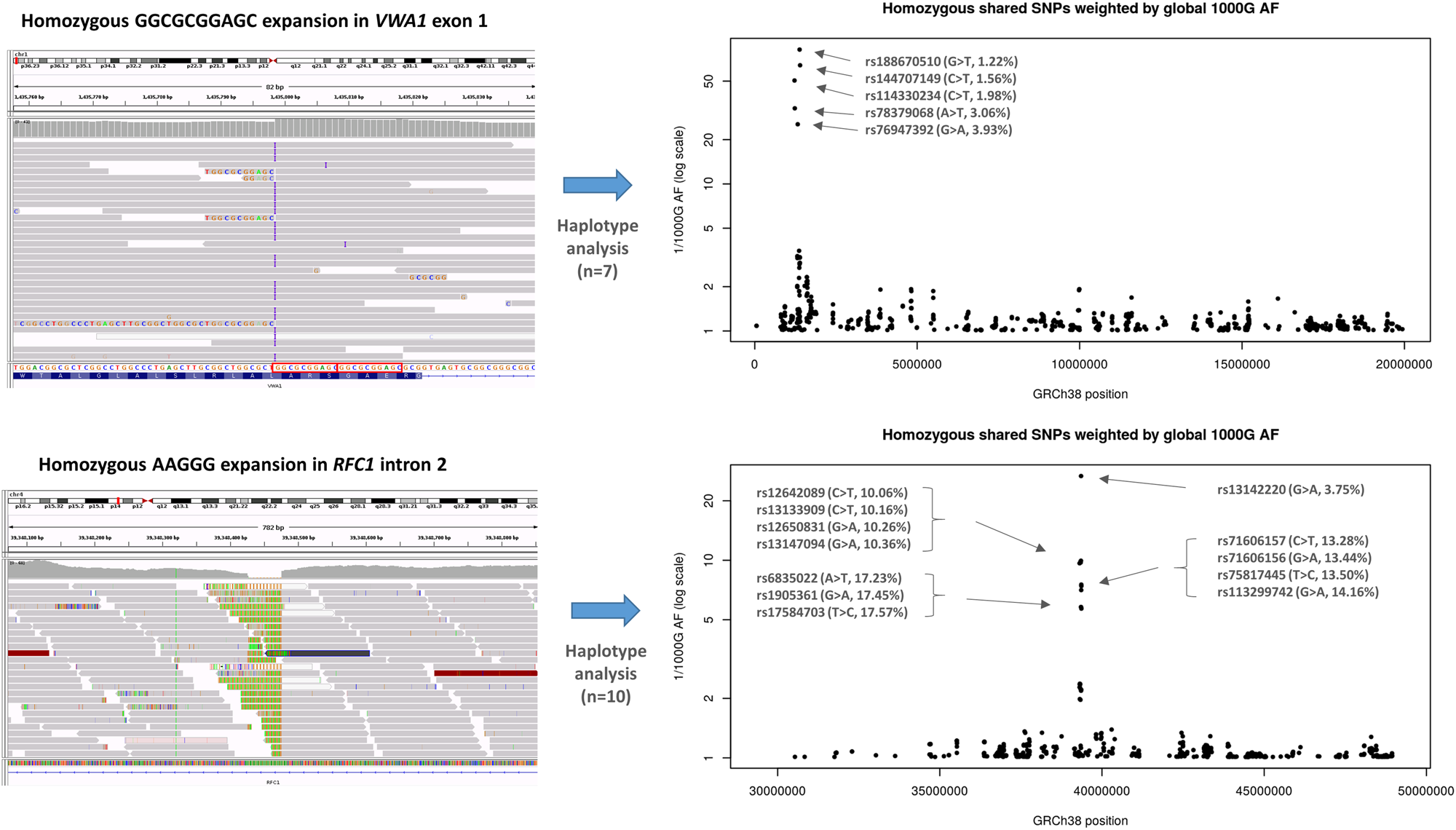
Fig. 2 Haplotype analysis performed on individuals with homozygous repeat expansions in VWA1 (top) compared to similar results for RFC1 (bottom). Shared homozygous SNPs are plotted for 20 Mb segments of chromosome 1 and 4. In both cases, the most significant regions detected span the VWA1 and RFC1 loci where several consecutive SNPs are homozygous in 7/7 or 10/10 individuals respectively. For the VWA1 locus, the five shared SNPs labelled with rsIDs are highly informative, with global 1000 Genomes project allele frequencies <5% (1000G AF). The reciprocal of the allele frequency is plotted on the y-axis to give an idea of the information content for each of the shared SNPs. Within the VWA1 haplotype block we also observed a rare SNV in the 5′-UTR of ANKRD65 (rs758603246), which is heterozygous in 3/7 individuals. The variants appear to be in complete linkage disequilibrium as in the 100KGP all individuals with rs758603246 also have p.G25Rfs*74. Our interpretation is that rs758603246 is a more recent mutation and so is observed only on a subset of p.G25Rfs*74-containing haplotypes.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Brain