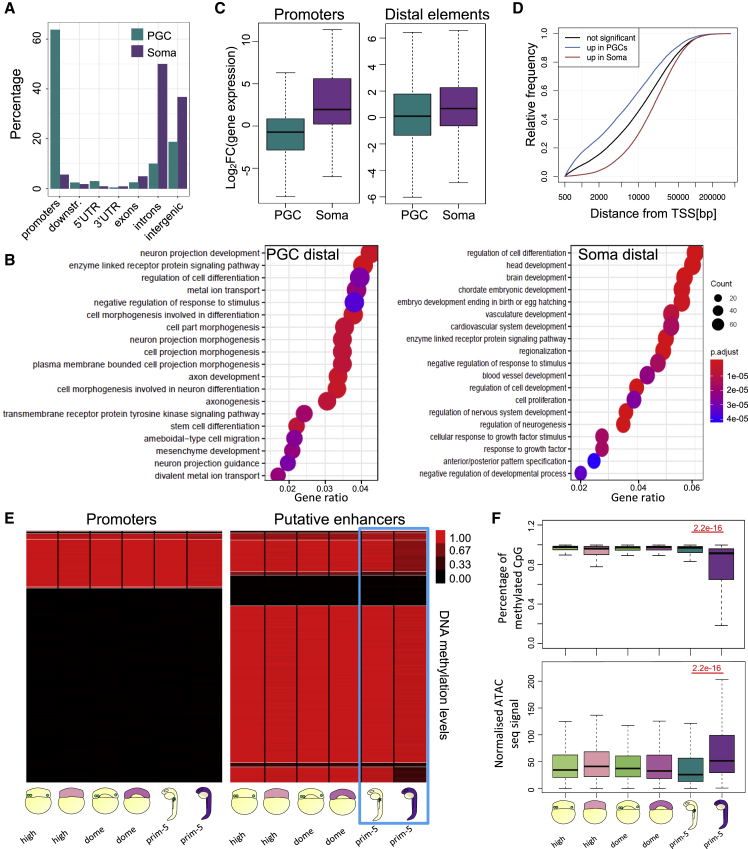
Figure 5 PGCs do not open chromatin at regions identified as putative enhancers (A) Percentage of differentially accessible ATAC peaks in PGCs and somatic cells at prim-5 stage overlapping with gene features. Promoter regions include 1 kb up- and downstream of the TSS. (B) Developmental processes GO analysis for genes associated with differentially accessible putative enhancers in PGCs and somatic cells. (C) Fold change of gene expression for genes associated with ATAC peaks upregulated in PGCs or somatic cells. Fold changes represent gene upregulation in PGCs and somatic cells, respectively. (D) Cumulative frequency of open chromatin elements in relation to distance from the closest TSS. padj < 0.05. Colors indicate elements near differentially expressed genes as indicated. (E) Heatmap of average DNA methylation levels at promoters (left) and H3K4me1/H3K27ac-rich genomic sites (putative enhancers) in PGCs and somatic cells at high, dome, and prim-5 stages. Blue box highlights prim-5 stage. (F) Quantification of methylated CpGs and chromatin accessibility at putative enhancer regions. p value in red was calculated by Wilcoxon test. Outliers are omitted.
Reprinted from Developmental Cell, 56(5), D'Orazio, F.M., Balwierz, P.J., González, A.J., Guo, Y., Hernández-Rodríguez, B., Wheatley, L., Jasiulewicz, A., Hadzhiev, Y., Vaquerizas, J.M., Cairns, B., Lenhard, B., Müller, F., Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization, 641-656.e5, Copyright (2021) with permission from Elsevier. Full text @ Dev. Cell