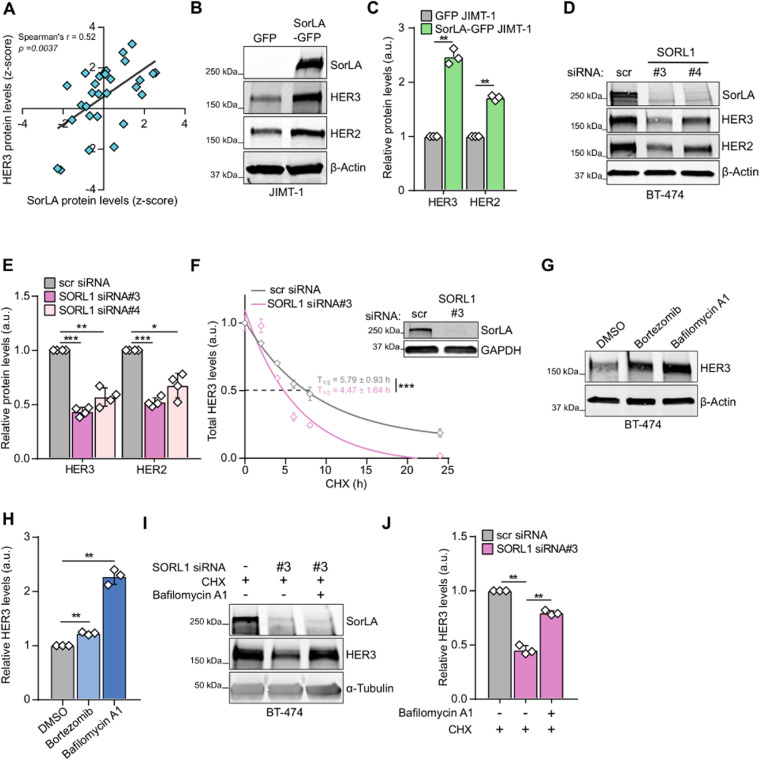
Fig. 3 A SorLA and HER3 protein levels correlate positively in breast cancer cell lines (DepMap portal; N = 29). B–E HER3 and HER2 expression correlates with SorLA in HER2-positive breast cancer. B, C SorLA-GFP transfection in JIMT-1 cells increases HER2 and HER3 levels compared to GFP transfected cells. D, E. SorLA silencing in BT-474 cells decreases HER2 and HER3. B, D Representative immunoblotting of total HER2, HER3, and SorLA with β-actin as a loading control. C, E represent the respective quantifications of immunoblots in (B, D) with HER2/HER3 levels normalized to loading control and relative to control-silenced cells. F SorLA silencing decreases HER3 stability. RNAi transfected BT-474 cells were treated with 25 µg.mL−1 of CHX for the indicated time points and HER3 protein levels were determined by immunoblotting (see Fig. S3E). Shown are HER3 levels normalized to α-tubulin and relative to 0 h timepoint. Half-lives (T1/2) represent the time required for HER3 to decrease to 50% of its initial level. The least squares fitting method and extra-sum-of-squares F test were used to assess the statistical difference between curves from control and SorLA-silenced cells (P = 0.0002). A representative western blot validating SorLA silencing is shown. G HER3 is primarily degraded through the lysosomal pathway. BT-474 cells were treated with 1 µM of bortezomib or 50 nM of bafilomycin A1 for 4 h to inhibit proteasome and lysosome activities, respectively. HER3 expression was analyzed by immunoblotting, with α-tubulin as a loading control. H Quantification of HER3 levels normalized to loading control and relative to DMSO-treated control cells. I SorLA silencing triggers HER3 lysosomal degradation. SorLA-silenced BT-474 cells were cotreated for 4 h with CHX and bafilomycin A1. HER3 expression was analyzed by immunoblotting, with α-tubulin as a loading control. J Quantification of HER3 levels normalized to loading control and relative to CHX-treated control-silenced cells. Data are mean ± SD from three (C, F, H, J) or four (E) independent biological replicates. Statistical analyses: Student’s t test (unpaired, two-tailed, unequal variance) unless indicated otherwise. Scr: control non-targeting siRNA.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Oncogene