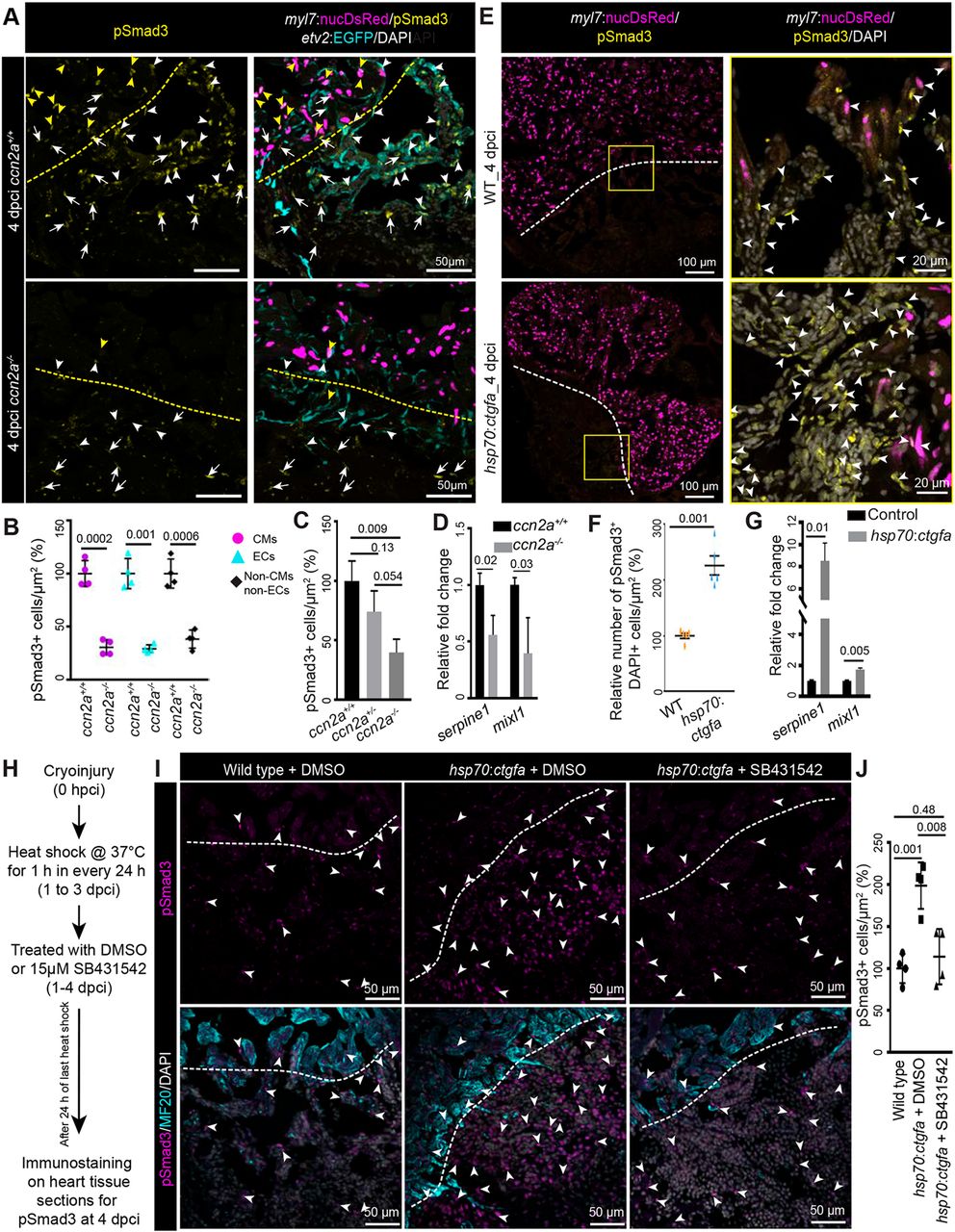
Fig. 7 Ccn2a levels positively modulate nuclear pSmad3 localization in injured hearts. (A) Maximum intensity projections of confocal images of sagittal cryosections of 4 dpci hearts expressing DsRed in CM nuclei (magenta) and EGFP in endocardial/endothelial cells (cyan), immunostained for pSmad3 (yellow) and stained with DAPI (white; marks all nuclei). Yellow and white arrowheads indicate DsRed+/pSmad3+ and EGFP+/pSmad3+ cells, respectively. White arrows indicate cells that are DAPI+/pSmad3+ but DsRed− and EGFP−. Dotted lines indicate the wound edge. (B) Percentage of Tgfβ activity quantified in ccn2a+/+ and ccn2a−/− hearts (n=4). The graph represents the ratio of nuclear DsRed+/pSmad3+ CMs to the total number of nuclear DsRed+ CMs in the border zone (CMs); the ratio of EGFP+/pSmad3+ cells to the total number of EGFP+ endocardial/endothelial cells in the injured tissue (ECs); and the ratio of pSmad3+/DAPI+ cells to the total number of EGFP- and DsRed- cells per unit area in the injured tissue (non-CMs and non-ECs). The mean wild-type control value was set to 100%. Quantifications were performed on two sagittal sections from each heart. (C) Percentage of Tgfβ activity quantified in ccn2a+/+, ccn2a+/− and ccn2a−/− hearts at 4 dpci (n=3). The graph represents the ratio of pSmad3+/DAPI+ nuclei to the total number DAPI+ nuclei in the injured tissue. The mean wild-type control value was set to 100%. Quantifications were performed on two sagittal sections from each heart. (D) qPCR analysis of serpine1 and mixl1 expression at 4 dpci (n=3, each sample is a pool of six hearts). (E) Maximum intensity projection of confocal images of sagittal cryosection through 4 dpci wild-type and ccn2a-overexpressing (hsp70:ctgfa) heart expressing DsRed in CM nuclei (magenta), immunostained for pSmad3 (yellow) and stained with DAPI (white; marks all nuclei). Arrowheads indicate pSmad3+/DAPI+ cells. Dotted lines indicate the wound edge. (F) Nuclear pSmad3+ cells in the injured tissue quantified in wild-type and ccn2a-overexpressing (hsp70:ctgfa) heart (n=5 each). Two sagittal heart sections from each heart were considered for quantification. The mean wild-type control value was set to 100%. (G) qPCR analysis of serpine1 and mixl1 expression at 4 dpci (n=3, each sample is a pool of four hearts). (H) Schematic of the experimental procedures used in I and J. (I) Maximum intensity projection of optical sections of sagittal cryosections through 4 dpci wild-type heart and DMSO or SB431542 treated ccn2a-overexpressing (hsp70:ctgfa) heart, immunostained for pSmad3 (magenta) and with MF20 (cyan), and stained with DAPI (white; marks all nuclei). Arrowheads indicate pSmad3+/DAPI+ cells. Dotted lines indicate the wound edge. (J) Percentage of Tgfβ activity quantified in wild-type, and in DMSO or SB431542-treated ccn2a-overexpressing hearts at 4 dpci (n=4). The graph represents the ratio of pSmad3+/DAPI+ nuclei to the total number DAPI+ nuclei in the injured tissue. The mean wild-type control value was set to 100%. Quantifications were performed on two sagittal sections from each heart. Data are mean±s.d. A two-tailed Student's t-test was used to evaluate the statistical significance of the differences (GraphPad Prism). Thickness of the each maximum projection is 10-12 µm. Mean Ct values are provided in Table S4.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development