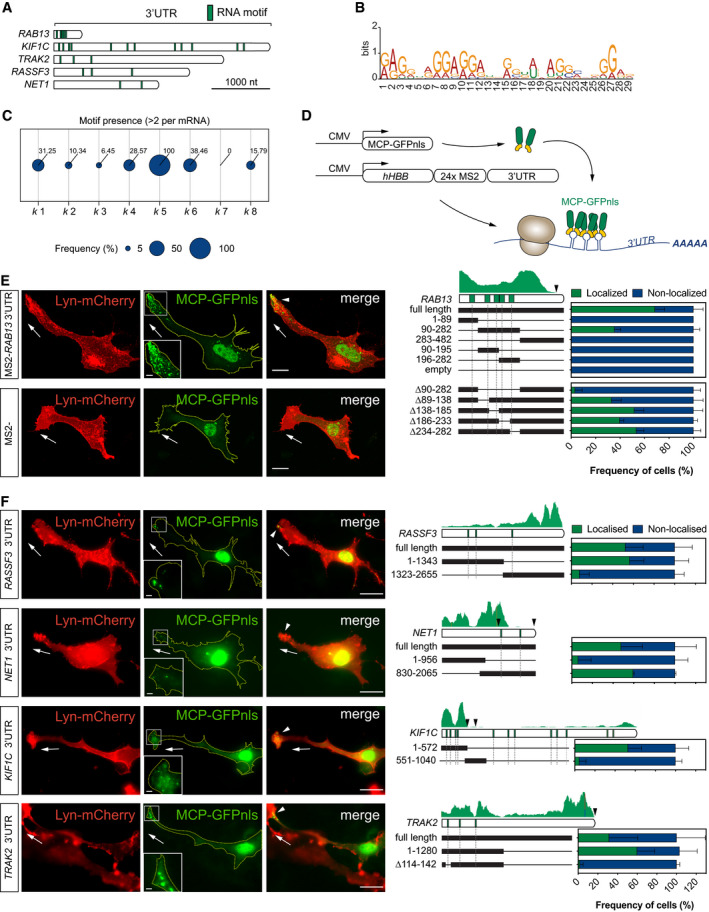
Figure 2
Diagram of RNA motif over‐represented in Frequency of mRNAs within each Scheme depicts the Left: representative subconfluent motile cells co‐transfected with plasmids expressing Lyn‐mCherry, MCP‐GFPnls and 24xMS2‐ Left: representative cells co‐transfected with plasmids expressing Lyn‐mCherry, MCP‐GFPnls and 24xMS2‐
Data information: white arrowheads indicate non‐nuclear localisation of MCP‐GFPnls; arrows indicate the orientation of RNA localisation; yellow dashed lines outline cell borders; scale bars = 20 μm (E, F); scale bars in insets = 5 μm (E) and 2 μm (F). For each