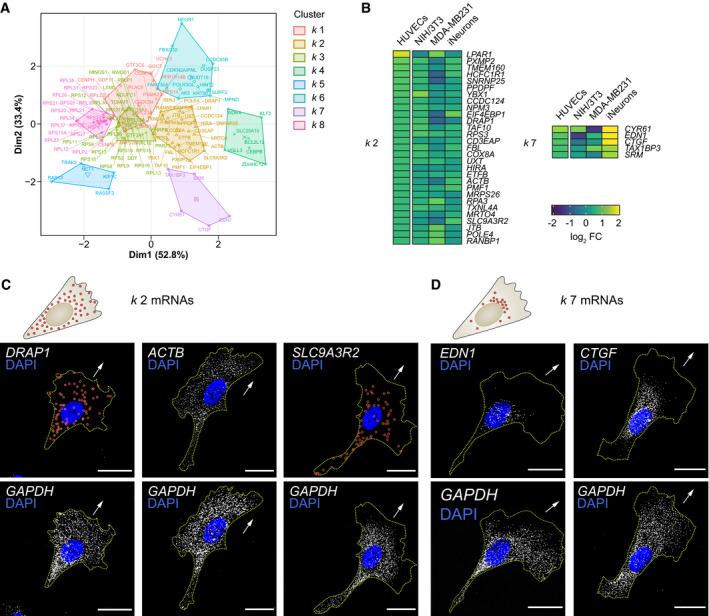
Figure EV1
Principal component plot depicting the Detail of the heat map shown in Fig Top: distribution pattern of mRNAs clustered in Top: distribution pattern of mRNAs clustered in
Data information: arrows indicate the orientation of RNA localisation; yellow dashed lines outline cell borders; red circles highlight smFISH spots; scale bars = 20 μm (C, D).