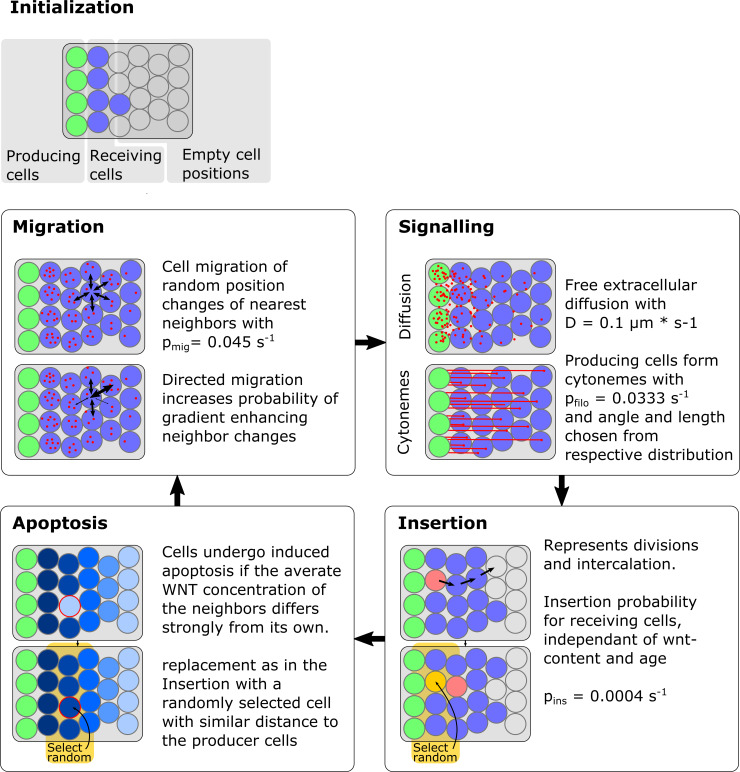
Fig 8
The field in which the tissue is modeled consists of precomputed non-overlapping potential cell positions (“fixed irregular lattice sites”). To initialize a simulation one margin of the field is filled with non-dividing morphogen producing cells (here shown in green), as well as morphogen receiving cells (shown in blue) with no initial morphogen concentration. After initialization (up to) four processes are sequentially executed for one timestep. In the