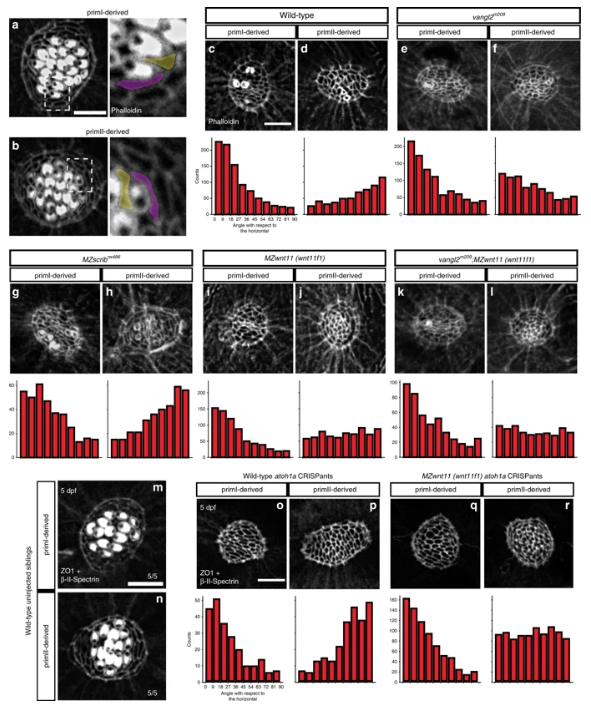
Fig. 5
Wnt pathway genes are required for proper coordinated support cell organization. a, b Phalloidin staining of wild-type primI and primII-derived neuromasts. The dotted square outlines the area magnified in (a) and (b). Two support cells each were false colored to depict their orientation. b–j Phalloidin staining of neuromasts 3 h after hair cell ablation. c, d Wild-type primI (c) and primII-derived (d) neuromasts. e, f vangl2 mutant primI (e) and primII-derived (f) neuromasts. g, h MZscrib mutant primI (g) and primII-derived (h) neuromasts. i, j MZwnt11 (wnt11f1) mutant primI (i) and primII-derived (j) neuromasts. k, l vangl2;MZwnt11 (wnt11f1) double mutant primI (k) and primII-derived (l) neuromasts. The histograms show the distribution of binned angles of cell orientation with respect to the horizontal for each of the conditions tested. c, d Angle distribution in wild-type primI (c one-way chi square test p = 1.78 × 10−87) and primII-derived (d p = 4.54 × 10−19) neuromasts. Note that primI distribution is skewed toward being horizontally aligned, while the distribution in primII-derived neuromasts is toward vertical alignment. e, f Angle distribution in vangl2 mutant primI (e p = 4.91 × 10−13) and primII-derived (f p = 2.14 × 10−16) neuromasts. g, h Angle distribution in MZscrib mutant primI (g p = 1.40 × 10−14) and primII-derived (h p = 9.60 × 10−13) neuromasts. i, j Support cell orientation in MZwnt11 (wnt11f1) mutant primI (i p = 5.32 × 10−52) and primII-derived (j p = 0.006) neuromasts. k, l Support cell orientation in vangl2;MZwnt11 (wnt11f1) double mutant primI- (k p = 6.69 x 10^-26) and primII-derived (l p = 0.22) neuromasts. m, n Double β-II-Spectrin (labeling the cuticular plates) and ZO-1 staining of a 5 dpf wild-type neuromasts. o, p Wild-type atoh1a CRISPants do not possess hair cells (β-II-spectrin is absent; see also Supplementary Fig. 5) and the support cells in all neuromasts show coordinated orientation (one-way chi-square test p-value in o = 4.98 × 10−20, p = 2.63 × 10−19). q, r MZwnt11 (wnt11f1) injected with atoh1a CRISPR show coordinated support cell orientation in primI-derived neuromasts (q p = 2.88 × 10−60) and do not show coordinated support cell orientation in primII neuromasts (r p = 0.24). Scale bar in a, c, and o equal 5 μm