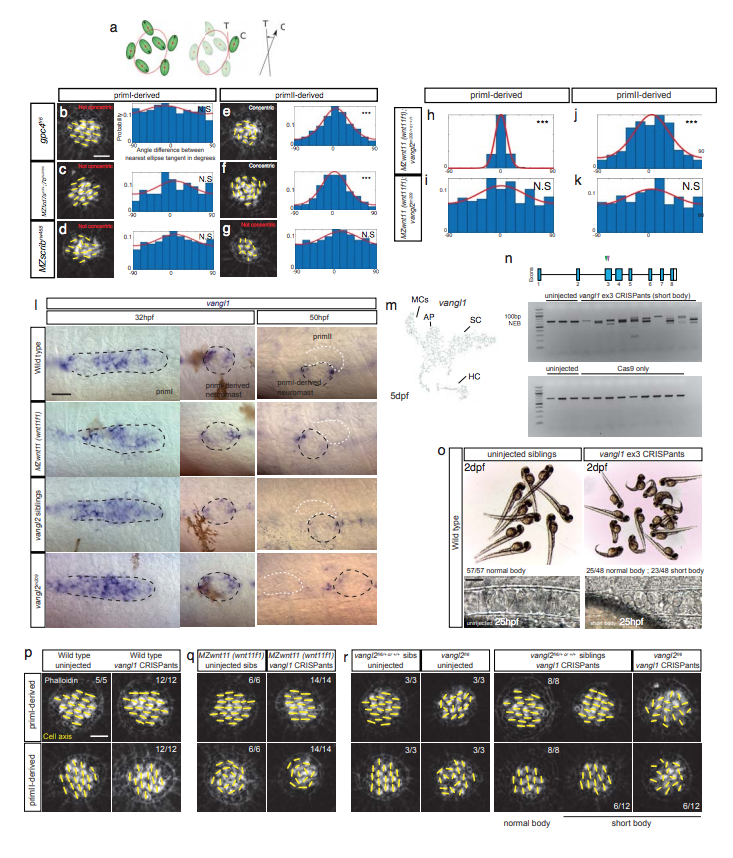
Fig. s2 Wnt and PCP pathway mutants show different hair cell phenotypes, but not due to redundancy with vang/1. a. Diagram showing how the analysis of angle difference between a given hair cell’s orientation and the nearest fitting ellipse (concentricity) in each of the conditions was calculated. The angle between a cell’s axis (C) with respect to the tangent (T) of the nearest fitted ellipse is calculated for each cell. For more details, see Materials and Methods. b-d. Phalloidin images showing the cell polarity axis (yellow lines) in priml-derived neuromasts of gpc4 (b), MZfzd7a/7b (c) and MZscrib mutants (d). Based on the distribution of angles with respect to the nearest ellipse tangent {concentricity) none of the mutants shows significant concentricity (Uniform distribution p-values in b= 0.9S, c= 0.666, d=0.16S; gpc4 n=222 hair cells, MZfzd7a/7b n=74, MZscrib n=535). Additionally, the von Mises distribution is shown in red for visual display of the data distribution. e-g. Phalloidin images showing the cell polarity axis (yellow lines) in primll-derived neuromasts of gpc4 (e), MZfzd7a/7b (f) and MZscrib (g) mutants. Only the Wnt pathway mutants show significant concentricity (Uniform distribution p-values in e= 2.03 x 10^-11, f= 2.0S x 10^-12, g=0.488; gpc4 n=214 hair cells, MZfzd7a/7b n=182, MZscrib n=392). Yellow lines in b-g indicate the hair cell polarity axis, determined by the position of the kinocilium. Not Concentric vs Concentric labels were based on statistical significance. h-k. Analysis of hair cell orientation with respect to the nearest neighbor. h-i. Angle distribution in priml-derived neuromasts of single MZwnt11 (wnt11f1) siblings {h) and MZwnt11 (wnt11f1);vang/2 mutants (i). The high degree of alignment in h (Uniform distribution p-value= 1.82 x 10^-223) is lost in i (Uniform distribution p-value= 0.039S). j-k. Angle distribution in primll-derived neuromasts of single MZwnt11 (wnt11f1) siblings (j) and double MZwnt11 (wnt11f1);vang/2 mutants (k). Note how the high degree of alignment in j (Uniform distribution p-value= 1.29 x 10^-14) is lost in k (Uniform distribution p-value= 0.069). l. vang/1 in situ in wild type, MZwnt11 (wnt11f1) mutants and vang/2 siblings and mutants. No evident changes in expression were observed. m. scRNAseq t-SNE plot for vang/1 expression in a 5dpf wild type neuromast, where its expression is low in mantle cells (MCs). n. Agarose gels showing the efficiency of CRlSPR targeting of exon 3 in vang/1 in CRlSPants (upper part) and Cas9-only injections (lower part). o. Brightfield images of 2dpf wild type uninjected siblings (left) and vang/1 CRlSPants (right), which show shortened, curly tails. Bottom panels: DlC images of 25 hpf live embryos showing the morphology of the notochord cells that are stacked in uninjected fish and are rounded in vang/1 CRlSPants, a characteristic of PCP defects. p-r. Phalloidin staining of the hair cell orientation in uninjected and vang/1 CRlSPR injected wild type (p), MZwnt11 (wnt11f1) (q) and vang/2 mutants (r). vang/1 targeting by CRlSPR does not affect hair cell orientation in wild type fish or modifies the phenotype characteristic of MZwnt11 (wnt11f1) and vang/2 mutants. Scale bar in b and p equals 5µm; l and o equals 20µm.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Commun.