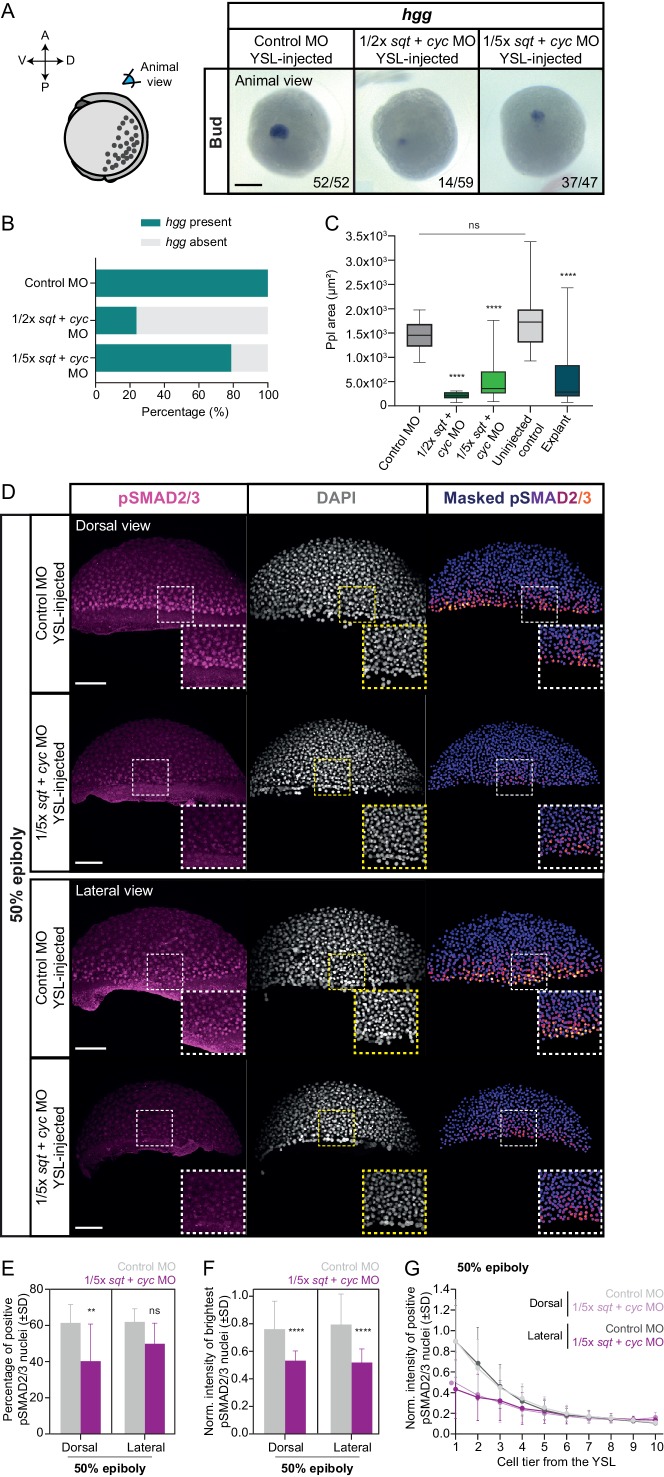
Figure 6 (A) Expression of hgg, a head mesoderm marker gene, in bud stage control MO (6 ng) or varying dosages of sqt + cyc MO YSL-injected embryos (1/2x: 1 ng sqt + 2 ng cyc MO and 1/5x: 0.4 ng sqt + 0.8 ng cyc MO), as determined by whole mount in situ hybridization. All embryos are shown as an animal view (schematic representation in the left). The proportion of embryos with a phenotype similar to the images shown is indicated in the lower right corner (control MO: n = 52, N = 3; 1/2x sqt/cyc: n = 59, N = 3; 1/5x sqt/cyc: n = 47, N = 3). (B) Percentage of control MO (n = 52, N = 3) or sqt + cyc MO YSL-injected embryos (1/2x sqt/cyc: n = 59, N = 3; 1/5x sqt/cyc: n = 47, N = 3) showing expression of hgg as determined by whole mount in situ hybridization at bud stage. (C) Ppl area (based on hgg staining, as determined by whole mount in situ hybridization) in control MO (n = 52, N = 3) or sqt + cyc MO YSL-injected embryos (1/2x sqt/cyc: n = 14, N = 3; 1/5x sqt/cyc: n = 37, N = 3), uninjected wildtype embryos (n = 31, N = 4) or blastoderm explants, prepared from 256 c embryos (n = 20, N = 5). ****p<0.0001, ns, not significant. (Kruskal-Wallis test). (D) High-resolution fluorescence images of control MO (6 ng) or 1/5x sqt (0.4 ng) + cyc (0.8 ng) MO YSL-injected embryos stained both for pSMAD2/3 (pink) and DAPI (grey) at 50% epiboly (dorsal domain: control, n = 11, N = 5; 1/5x sqt/cyc, n = 8, N = 3; lateral domain: control, n = 9, N = 4; 1/5x sqt/cyc, n = 7, N = 3). Nuclear pSMAD2/3 is color-coded using a fire lookup table (highest intensities in yellow) and was masked based on the DAPI signal. Insets are zoom-in images of the highlighted regions (dashed boxes). (E) Percentage of pSMAD2/3 positive nuclei in control MO (6 ng) or 1/5x sqt (0.4 ng) + cyc (0.8 ng) MO YSL-injected embryos at 50% epiboly (dorsal domain: control, n = 11, N = 5; 1/5x sqt/cyc, n = 8, N = 3; lateral domain: control, n = 9, N = 4; 1/5x sqt/cyc, n = 7, N = 3). **p=0.0086, ns, not significant (Kruskal-Wallis test). (F) Normalized intensity of the brightest pSMAD2/3 nuclei (for details see Materials and methods) in control MO (6 ng) or 1/5x sqt (0.4 ng) + cyc (0.8 ng) MO YSL-injected embryos at 50% epiboly (dorsal domain: control, n = 11, N = 5; 1/5x sqt/cyc, n = 8, N = 3; lateral domain: control, n = 9, N = 4; 1/5x sqt/cyc, n = 7, N = 3). ****p<0.0001 (Kruskal-Wallis test). (G) Normalized intensity of pSMAD2/3 positive nuclei as a function of the distance to the YSL, expressed as cell tiers, in control MO (6 ng) or 1/5x sqt (0.4 ng) + cyc (0.8 ng) MO YSL-injected embryos at 50% epiboly (see Materials and methods for additional details; dorsal domain: control, n = 11, N = 5; 1/5x sqt/cyc, n = 8, N = 3; lateral domain: control, n = 9, N = 4; 1/5x sqt/cyc, n = 7, N = 3). (D-G) The position along the dorsal-ventral axis is indicated in the top right corner or at the bottom. Part of the control MO samples for (A-G) is also shown in Figure 5A and C–G. Scale bars: 200 µm (A), 100 µm (D).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife