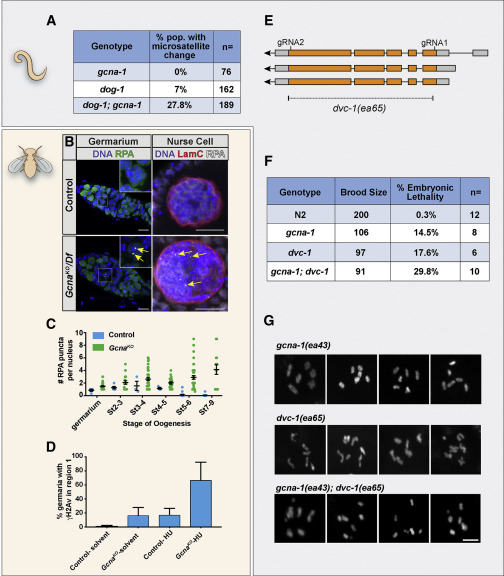
Fig. 3 GCNA Mutant Germ Cells Experience Replication Stress (A) Frequency of microsatellite repeat length changes within the C. elegans vab-1 locus. (B and C) RPA foci (yellow arrows) accumulate in Drosophila GcnaKO germ cells. (B) Control and GcnaKO/Df Germaria (left) and nurse cell nuclei (right) stained for DNA (blue), RPA (green), and Lamin C (red) as indicated. Scale bars represent 10 μm. (C) Quantification of RPA punctae per nucleus. 13-49 nuclei were counted per genotype per stage in triplicate. (D) Quantification of γH2Av foci in region 1 of the Drosophila germarium in control and GcnaKO mutant germaria in response to HU treatment. 37-52 germaria were counted per genotype per stage in triplicate. Mean values are shown with standard deviation. (E) The dvc-1/Spartan locus of C. elegans showing the strategy for creating a complete null, dvc-1(ea65). (F) Brood analysis (total viable adult offspring) and embryonic lethality associated with N2, dvc-1 and gcna-1 single mutants, and dvc-1;gcna-1 double mutant worms. n > 6 broods/genotype, 3-way Kruskal–Wallis test, p < 0.05. (G) Examples of DAPI-stained, diakinesis oocytes from dvc-1, gcna-1, and dvc-1; gcna-1 mutant germ cells show chromosome fragmentation in the double mutant. Scale bars represent 5 μm.
Reprinted from Developmental Cell, 52(1), Bhargava, V., Goldstein, C.D., Russell, L., Xu, L., Ahmed, M., Li, W., Casey, A., Servage, K., Kollipara, R., Picciarelli, Z., Kittler, R., Yatsenko, A., Carmell, M., Orth, K., Amatruda, J.F., Yanowitz, J.L., Buszczak, M., GCNA Preserves Genome Integrity and Fertility Across Species, 38-52.e10, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell