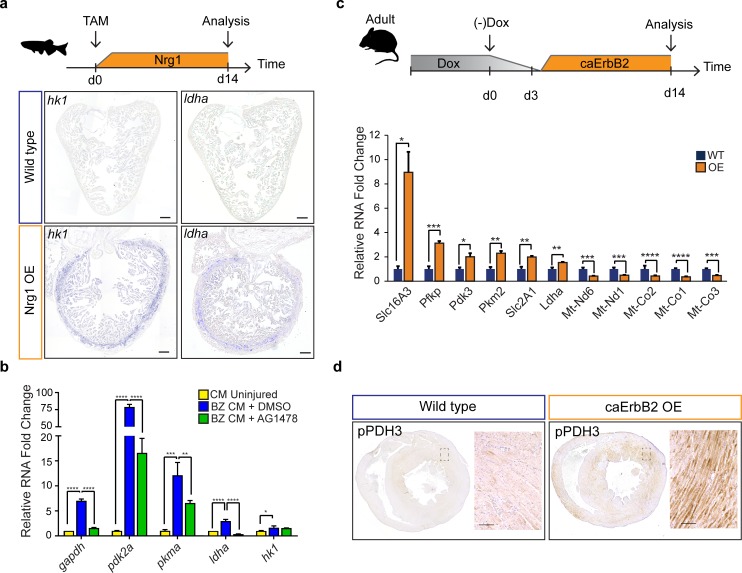
Figure 6 Nrg1/ErbB2 signaling induces glycolysis genes in zebrafish and mouse. (a) Cartoon showing experimental procedure to induce cardiomyocyte specific Nrg1 expression in zebrafish. Panels show in situ hybridization for hexokinase 1 (hk1) and lactate dehydrogenase a (ldha) expression on sections of control hearts (β-act2:BSNrg1) and Nrg1 OE hearts (cmlc2:CreER; β-act2:BSNrg1). Scale bars represent 100 μm. (b) qPCR results for glycolytic genes showing their relative fold change in DMSO treated (n = 9) (blue) and AG1478 treated (n = 9) (green) nppa:mCitrine high border zone cardiomyocytes at 3dpi compared to uninjured adult cardiomyocytes (n = 4) (yellow). Error bars represent standard deviation. (c) Upper panel: Cartoon showing the experimental procedure to analyse metabolic gene expression after activating ErbB2 signaling in the murine heart. Lower panel: qPCR results for metabolic genes showing their relative fold change in caErbB2 OE (n = 4) heart compared to control WT hearts (n = 4). Error bars represent standard deviation. (d) Immunohistochemistry for phospho-PDH3 on sections of control and caErbB2 OE hearts. Scale bars represent 100 μm. *=p < 0.05, **=p < 0.01, ***=p < 0.001, ****=p < 0.0001.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife