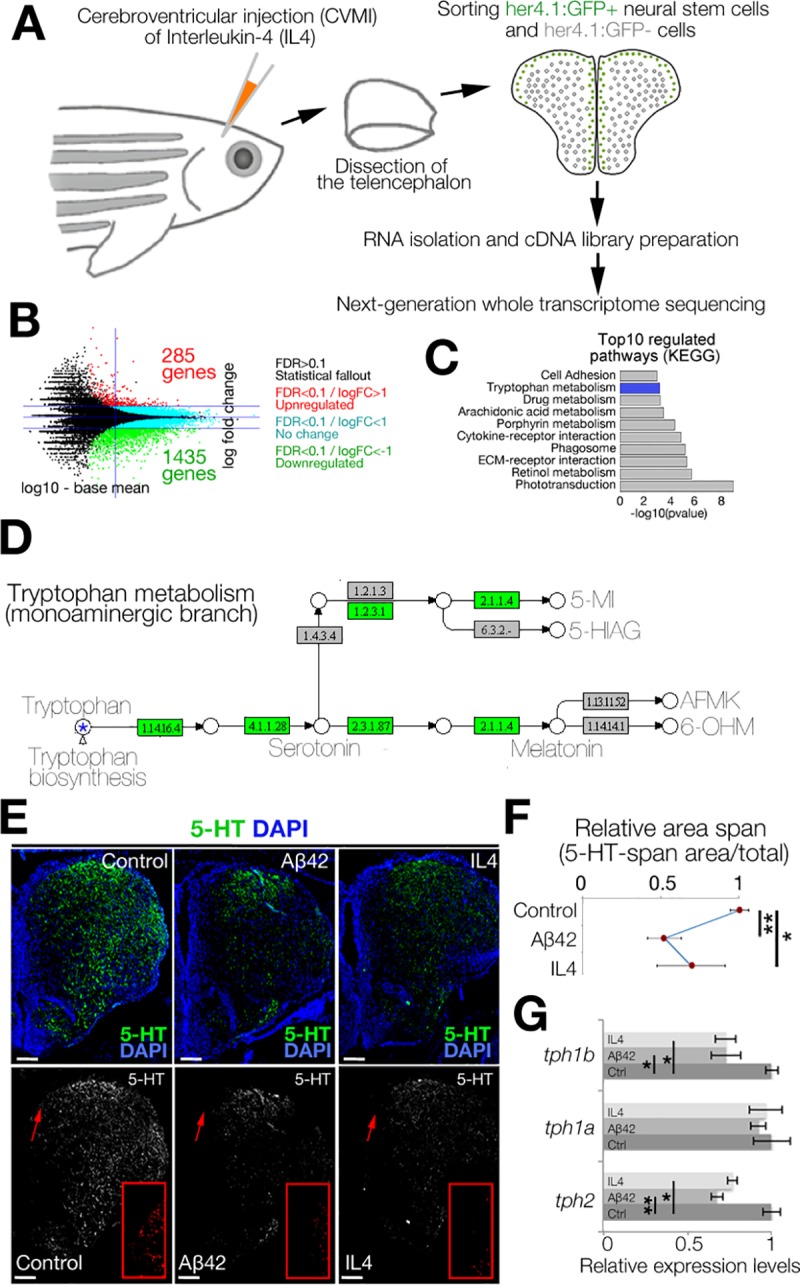
Fig 1
(A) Schematic view of the experimental pipeline for whole-transcriptome sequencing IL4 treatment. (B) MA-plot for DEGs. (C) GO-term analyses on DEGs. (D) Modified KEGG-pathway view of tryptophan metabolism. Green indicates the enzymes down-regulated by IL4. (E) IHC for 5-HT in control (left), Aβ42-injected (middle), and IL4-injected (right) brains. Single-channel images show 5-HT. Red insets are high-magnification images of arrowed regions. (Dm: dorsal-medial) (F) Quantification of 5-HT-span area density under the conditions of E. (G) qRT-PCR results for