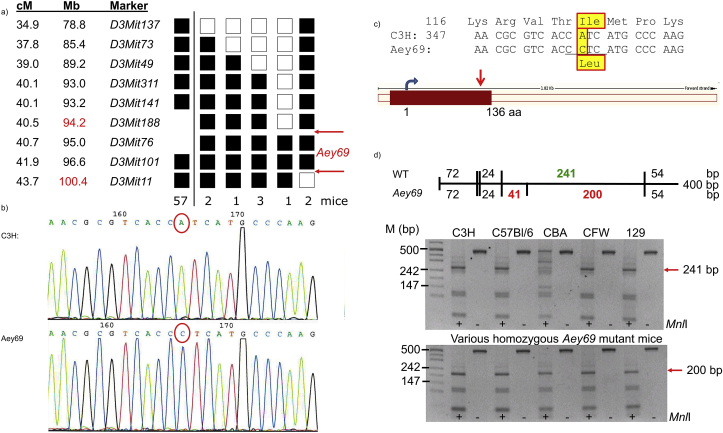
Fig. 3
Reprinted from Experimental Eye Research, 188, Vetrivel, S., Tiso, N., Kügler, A., Irmler, M., Horsch, M., Beckers, J., Hladik, D., Giesert, F., Gailus-Durner, V., Fuchs, H., Sabrautzki, S., German Mouse Clinic, Helmholtz Zentrum München, German Research Center for Environmental Health GmbH, Neuherberg, Germany, Adler, T., Treise, I., Busch, D.H., Aguilar-Pimentel, A., Ollert, M., Götz, A., Amarie, O.V., Stoeger, T., Schulz, H., Becker, L., Klopstock, T., Schrewe, A., Spielmann, N., Bekeredjian, R., Garrett, L., Hölter, S.M., Zimprich, A., Wurst, W., Mayer-Kuckuk, P., Hans, W., Rozman, J., Klingenspor, M., Neff, F., da Silva-Buttkus, P., Calzada-Wack, J., Rácz, I., Zimmer, A., Rathkolb, B., Wolf, E., Prehn, C., Adamski, J., Östereicher, M., Miller, G., Steinkamp, R., Lengger, C., Maier, H., Stoeger, C., Leuchtenberger, S., Gailus-Durner, V., Fuchs, H., Hrabě de Angelis, M., Hrabě de Angelis, M., Graw, J., Mutation in the mouse histone gene Hist2h3c1 leads to degeneration of the lens vesicle and severe microphthalmia, 107632, Copyright (2019) with permission from Elsevier. Full text @ Exp. Eye. Res.