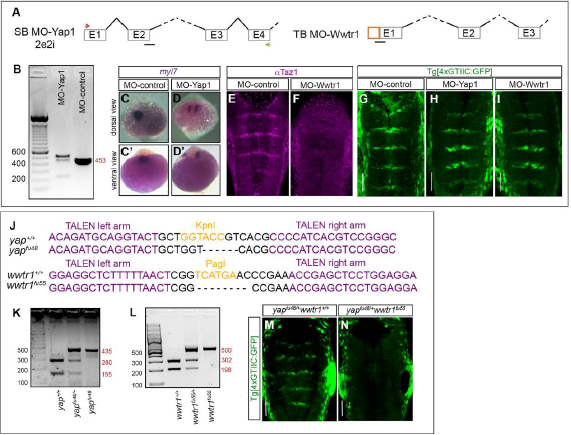
Fig. S3
Loss-of-function of Yap and Taz: morpholinos and genomic edition of yap and wwtr1 genes by TALEN technology.
A) Scheme depicting the structure of the different employed mopholinos. MO-Yap: position of the splice-blocking morpholino (MO-Yap1SB2e2i, black line), and the primers used to assess its efficiency (see green and red arrows). MO-Wwtr1 is a translation-blocking morpholino for Taz. B) Agarose gel showing the aberrant splicing products induced by MO-Yap as detected by RT-PCR of embryos injected with either MO-control or MO-Yap1. C-D) Embryos injected with either MO-control or MO-Yap1 and in situ hybridized for myl7. Note that upon downregulation of Yap1, embryos displayed defects in the migration of heart progenitors (compare C-C’ with D-D’) as previously described (Fukui et al., 2014). Whole embryo dorsal (C-D) or ventral (C’-D’) views with anterior to the top. E-F) Embryos injected with either MO-control or MO-Wwtr1 and immunostained with anti-Taz. Note that Taz protein expression is downregulated in the MO-Wwtr1 injected embryos demonstrating that MO-Wwtr1 does indeed interfere with the translation of Taz. G-I) Analysis of Yap/Taz-TEAD activity in embryos where Yap or Taz was downregulated using morpholinos (MO-control: n = 16; MO-Yap: n = 23; MO-Wwtr1: n = 26). Note that no effects on Yap/Taz-TEAD activity were observed when either cofactor was downregulated alone. Embryos injected with MO-Yap and MO-Wwtr1 resulted in severe malformations and did not progress until the analysis stage. Images in (E-J) are hindbrain dorsal views with anterior to the top. J) Alignment of the yapfu48 and wwtr1fu55 mutant alleles with their corresponding wild-type (yap+/+ and wwtr1+/+) sequences showing: i) the deleted nucleotides, and ii) the TALEN target sites in the yap and wwtr1 loci; the left and right arms (magenta) of these sites are separated by a spacer (black) including the restriction site used for screening (orange). K-L) Agarose gels with the obtained bands resulting from enzyme restriction mapping of gDNA corresponding to embryos carrying the wild type or the mutated alleles. The numbers in red indicate the size of the different obtained fragments. M-N) yapfu48/+wwtr1+/+ and sibling yapfu48/+wwtr1fu55 compound mutants in the TEAD-activity reporter background. Note that embryos in which three of the yap/wwtr1 alleles were mutated (yapfu48/+wwtr1fu55) do not display Yap/Taz-TEAD activity in the hindbrain boundaries (n=12/14), in comparison with control embryos (yapfu48/+wwtr1+/+, n=14/14 with TEAD-activity). Double homozygous yapfu48wwtr1fu55 embryos displayed severe deformations and did not survive until the analysis stage. Scale bars correspond to 50m.