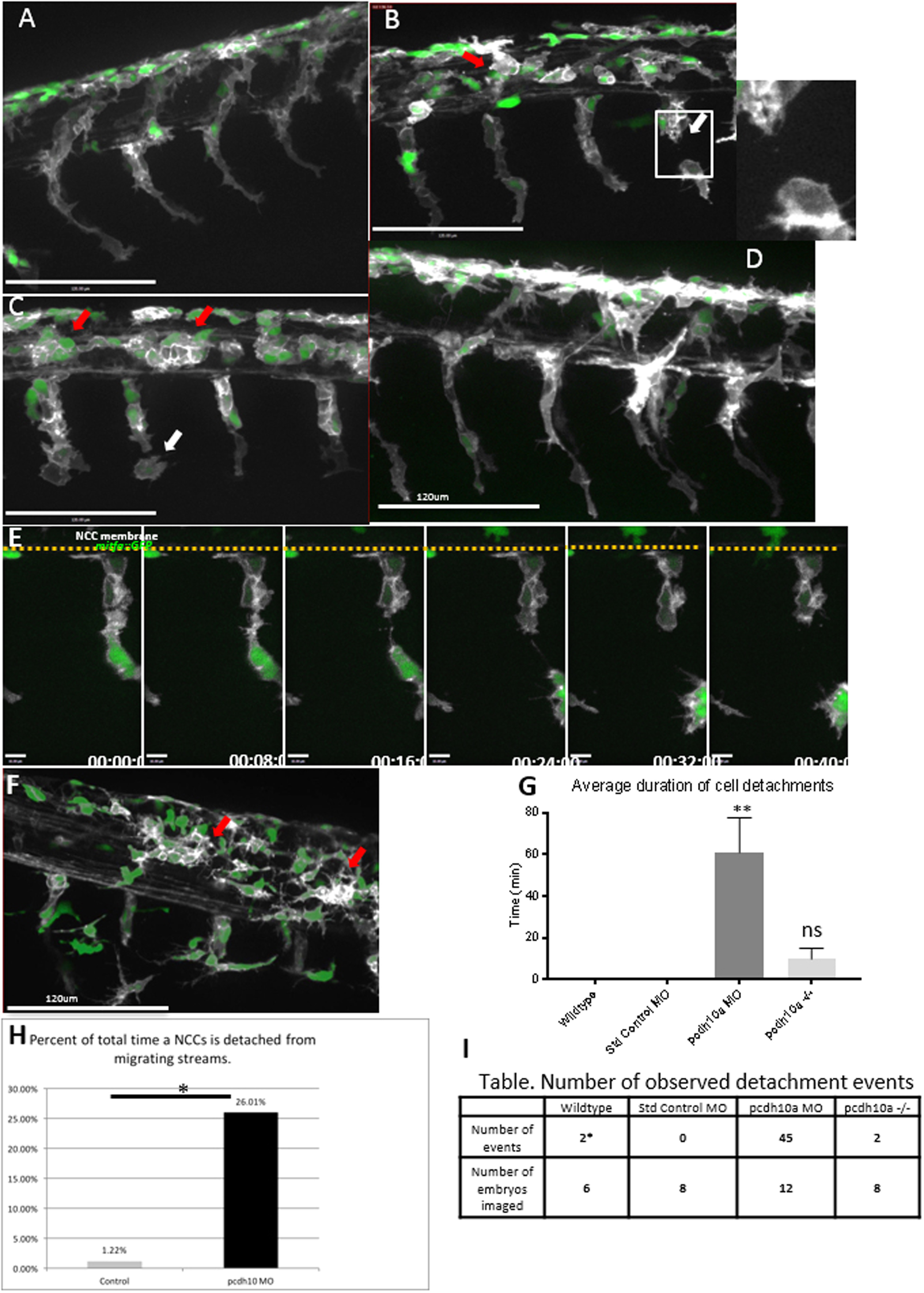
Fig. 4
pcdh10a knockdown causes defects in NCC migration.(A-D, F) Lateral views of double transgenic zebrafishTg(sox10:mRFP;mitfa:GFP), membrane RFP has been changed to gray scale. (A) Control non-injected NCCs 28hpf. (B,C) Lateral view of pcdh10a morphant NCCs at 28hpf show defects in NCC migration, including in mitfa+ melanoblasts. White arrows mark NCCs that have detached from other NCCs in the migrating streams. Red arrows mark NCCs that have aggregated within the neural tube domain. (D) Lateral view at 28hpf of embryos injected with Standard Control MO. Scale bars are 120 µm. (E) Time series showing the detachment of a neural crest cell in pcdh10a MO injected embryos, scale bars are 10 µm. (F) pcdh10a -/-Tg(sox10:mRFP;mitfa:GFP) embryo with subtle NCC aggregations (red arrows). (G) Quantification of the duration of cell detachment events across all samples. (H) Percent of total time NCCs are detached from other NCCs in Control and Morphant embryos.(I) Table, comparison of the number of embryos imaged and the number of detachment events that occurred across all samples. * denotes P value of 0.0062.
Reprinted from Developmental Biology, 444 Suppl 1, Williams, J.S., Hsu, J.Y., Rossi, C.C., Artinger, K.B., Requirement of zebrafish pcdh10a and pcdh10b in melanocyte precursor migration, S274-S286, Copyright (2018) with permission from Elsevier. Full text @ Dev. Biol.