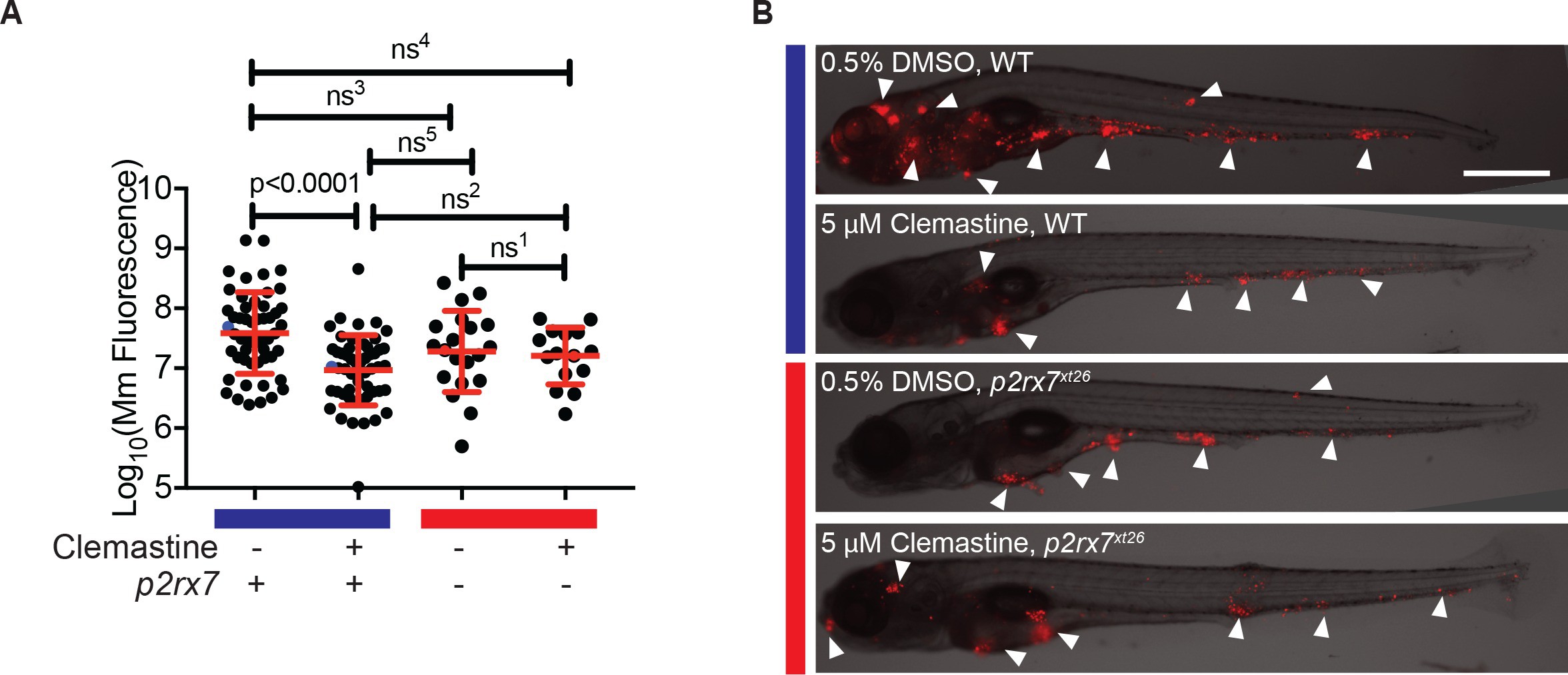
Fig. 4
Clemastine increases frequency of macrophage calcium transients in a p2rx7-dependent manner.
(A) Schematic of P2rx7 receptor, based on human structure, with CRISPR target site in exon 10 denoted with a star. (B) CRISPR/Cas9-mediated lesions in p2rx7 include a five base pair deletion (p2rx7xt26) and a two base pair deletion (p2rx7xt28) leading to a premature stop codon in exon 10. Red bar denotes p2rx7 mutants and blue bar denotes WT animals. (C) Quantification of calcium flashes observed with light-sheet microscopy in Tg(mfap4:GCaMP6F)xt25 and p2rx7xt26;Tg(mfap4:GCaMP6F)xt25 infected with ~ 50 CFU Mm:tdTomato. Each dot represents the number of flashes observed in a single infected macrophage in an animal during a 30-min time-course during treatment with 0.5% DMSO or 5 µM clemastine, 4–5 hpi, n > 3 fish for each group. All cells counted were visibly infected with Mm:tdTomato and scoring performed blind to genotype or treatment. (D) Representative calcium transient. Panels are stills from a light-sheet video in an untreated Tg(mfap4:GCaMP6F)xt25 animal infected with Mm:tdTomato. Scale bar is 25 µm. (C) Kruskal-Wallis ANOVA for unequal variances, Dunn’s multiple comparison test. ns1 > 0.9999, ns2 = 0.7007 All error bars are s.d.; p values from statistical tests on untransformed data are provided in Supplementary file 2.