Fig. 1
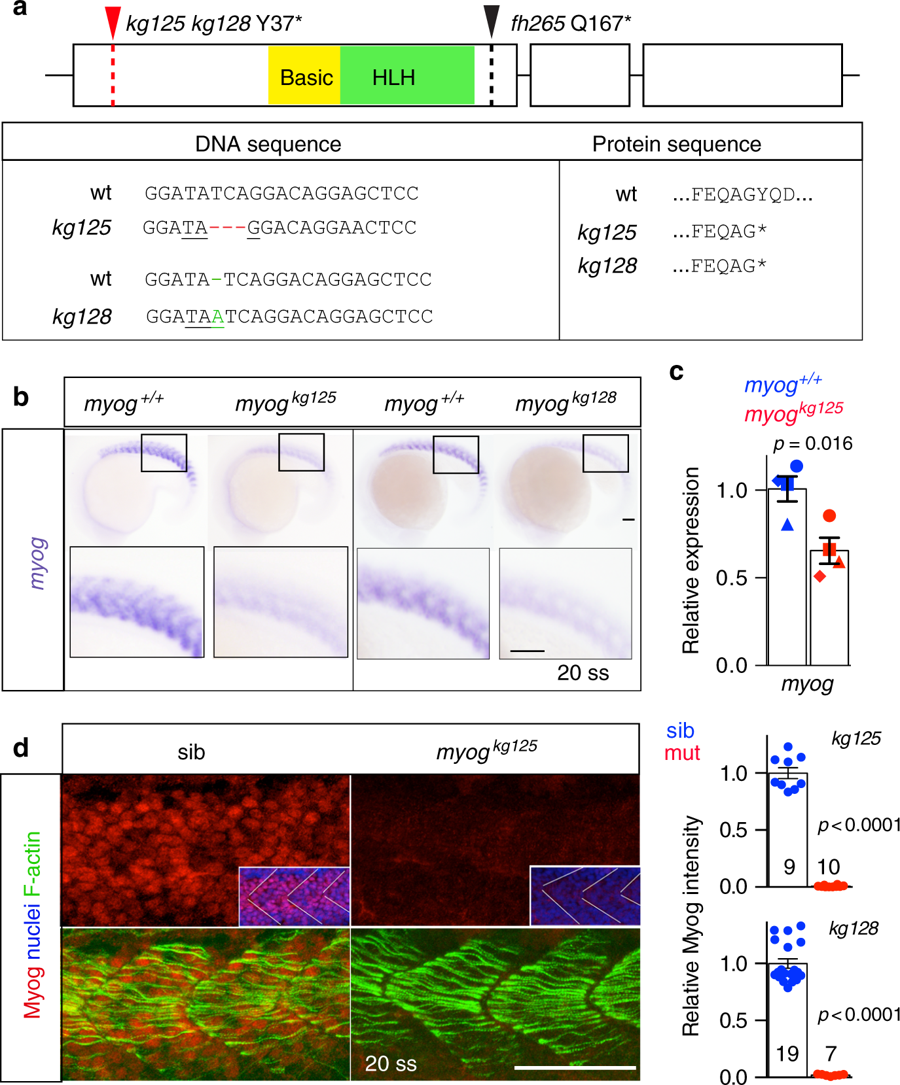
Genome editing generates zebrafish myog null alleles. a Schematic of myog gene exons (boxes) and protein showing the position of kg125, kg128 and fh265 mutant alleles. The tyrosine to stop mutations (Y37*) produce a truncated protein of 36 amino acids (aa) devoid of both basic (yellow) and helix–loop–helix (HLH, green) domains. The fh265 hypomorphic allele (Q167*) truncates downstream of bHLH. Beneath, DNA and protein alignment of wild-type (wt), 3 bp kg125 deletion (red) and 1 bp kg128 insertion (green) alleles with novel stop codons underlined causing an identical truncation. b In situ mRNA hybridisation (ISH) for myogenin on myogkg128/+ and myogkg125/+ incross lays reveal NMD of mutant myog mRNA at 20 somite stage (20 ss). Representative images n = 14 + 5 mutants, n = 39 + 22 sibs, respectively. Insets show magnification of boxed areas. c qPCR analysis on wt sibs and myogkg125 embryos at 22 ss confirms NMD. Mean fold change ± SEM from four independent experiments on genotyped embryos from four separate lays analysed on separate days, paired t test statistic. Symbol shapes denote matched wt and mutant samples from each experiment. d Immunoreactivity of Myog is lost in myogkg125 and myogkg128 mutants at 20 ss, whereas F-actin is unaffected. Insets show nuclear staining in myogkg125 and sib using Hoechst counterstain. Relative myotomal Myog immunofluorescence was assessed by nuclear intensity measurement. All images are lateral views anterior to left, dorsal to top. Representative images n = 10 + 7 mutants, n = 9 + 19 sibs, respectively. Bars = 50 µm