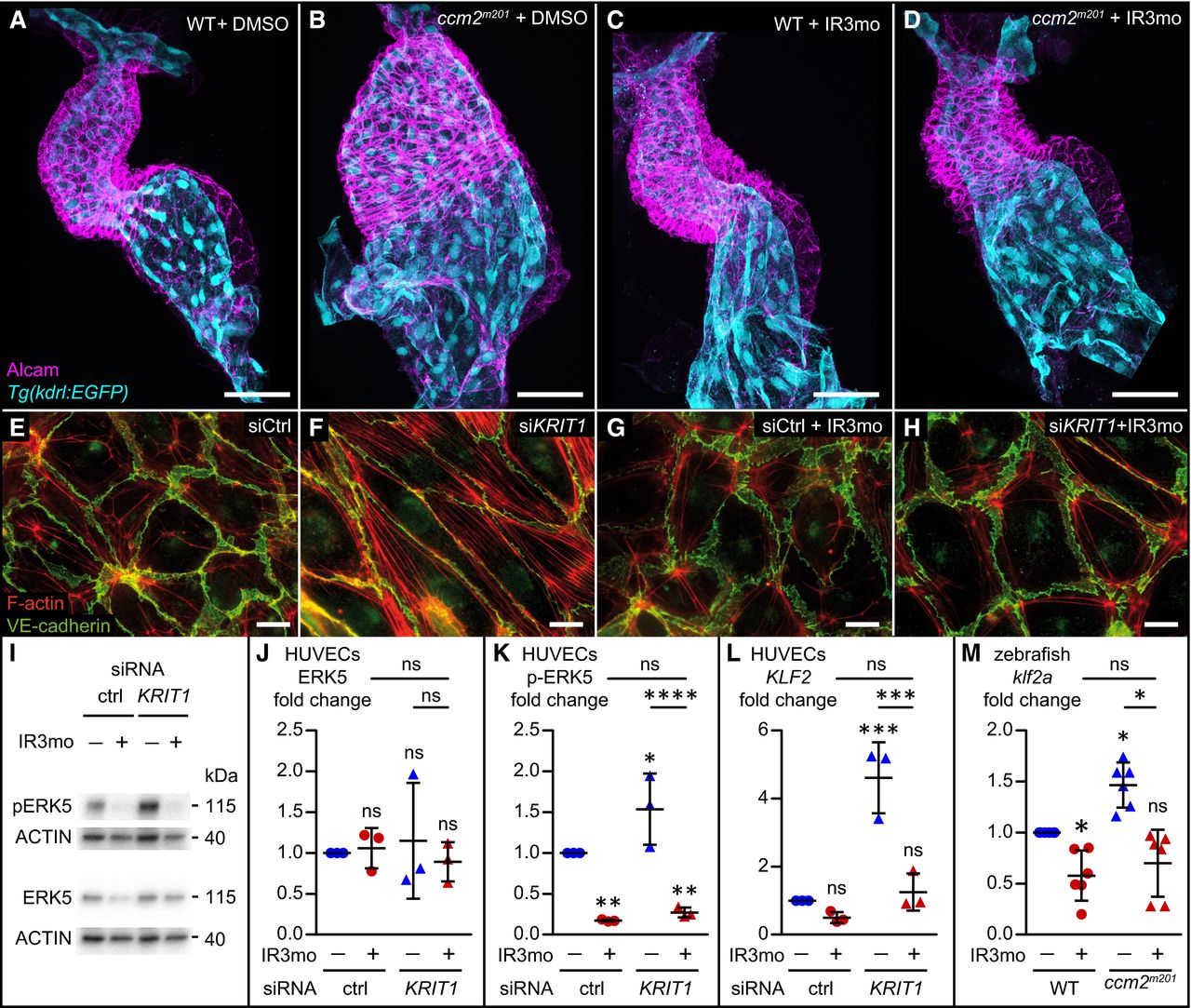
Fig. 4
Indirubin‐3‐monoxime (IR3mo) rescues the CCM phenotype in zebrafish and HUVEC models
A–D Treatment with 5 μM indirubin‐3‐monoxime (IR3mo) rescues the embryonic zebrafish ccm2m201 mutant ballooning heart phenotype. Shown are images of confocal z‐scan projections of wild‐type (WT) (A, C) and ccm2m201 mutant (B, D) zebrafish embryonic hearts at 48 hpf expressing the endothelial reporter Tg(kdrl:GFP)s843 (cyan) and counter‐stained for Alcam (magenta, labeling the myocardium). Scale bar is 50 μm.
E–H Treatment with 10 μM IR3mo for 48 h restores the wild‐type ACTIN‐adhesive phenotype in KRIT1‐depleted HUVECs. Shown are confocal images of HUVECs with labeled VE‐cadherin (green) and F‐ACTIN (red). Control siRNA‐silenced (E, G) and KRIT1 siRNA‐silenced (F, H) HUVECs were not treated (E, F) or treated (G, H) with IR3mo. Scale bar is 10 μm.
I–K Treatment with IR3mo reduces phosphorylation of ERK (pERK) without affecting overall ERK protein levels. Shown in (I) are representative Western blots of KRIT1‐silenced HUVECs lysates treated or not with 10 μM of IR3mo during transfection. ERK5 (J) and phosphorylated ERK5 protein levels (K) were measured relative to ACTIN protein levels based on three biological replicates with two technical replicates each (n = 3). Statistical analyses were performed using two‐way ANOVA followed by Tukey's multiple comparisons test; error bars are SD. ****P < 0.0001; ns, not significant.
L. Treatment with IR3mo rescues the elevated KLF2 levels of KRIT1‐silenced HUVECs. RT‐qPCRs were performed to measure KLF2 levels, on three biological replicates (n = 3). Statistical analyses were performed using one‐way ANOVA followed by Tukey's multiple comparisons test; error bars are SD. ***P < 0.001; ns, not significant.
M. Treatment with IR3mo rescues the elevated klf2a levels of ccm2m201 mutant zebrafish embryos at 48 hpf. RT‐qPCRs were performed to measure klf2a levels, on six biological replicates (n = 6). Statistical analyses were performed using one‐way ANOVA followed by Tukey's multiple comparisons test; error bars are SD. *P < 0.05; ns, not significant.