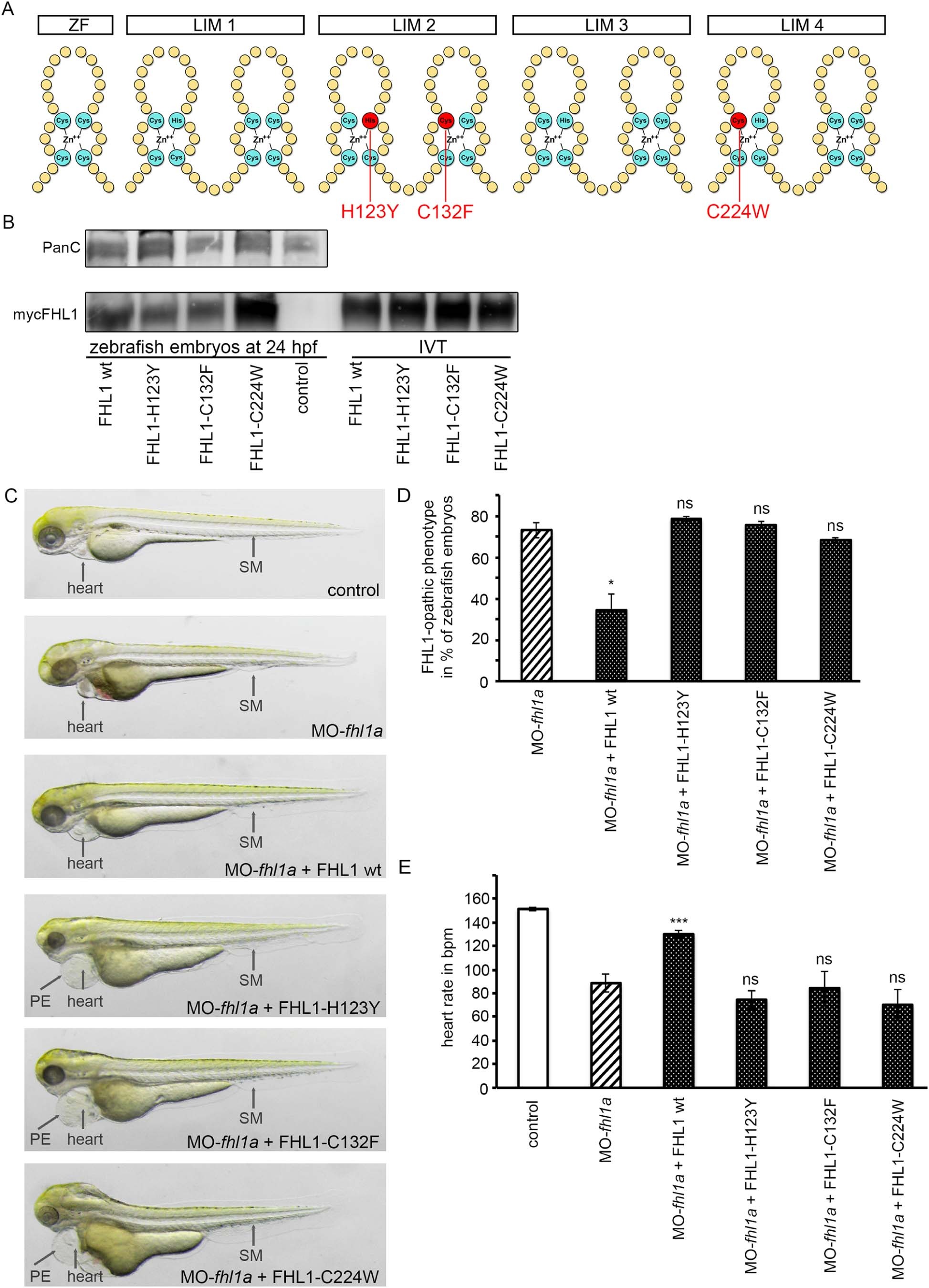
Fig. 4
(A) Schematic structure of FHL1. One Zinc finger domain (ZF) and four LIM domains (double Zinc finger motif, LIM1-LIM4). Three common human mutations are marked in red (H123Y, C132F and C224W). (B) Western blot of zebrafish embryos at 24 hpf (left) and in-vitro-translatate (IVT, right) staining, Pan Cadherin C as control and myc-expressing FHL1 with myc-antibody. mRNA of wild-type FHL1 and its mutated forms is translated into stable protein in zebrafish embryos and therefore can be detected in FHL1-mRNA injected zebrafish embryos but not in control embryos. (C) Phenotype of either control-injected or fhl1a-depleted zebrafish embryos. The lower four embryos are fhl1a-depleted and either injected with wild-type FHL1-mRNA or mutated FHL1-mRNA. When MO-fhl1a and wild-type (wt) FHL1-mRNA are co-injected, the phenotype resembles wild-type zebrafish. The skeletal muscle (SM) of MO-fhl1a and wild-type FHL1-mRNA co-injected embryos displays regular structural organization. Heart function is normalized (see Fig. 4E). In contrast to human wild-type FHL1, the mutated human FHL1-proteins (FHL1-H123Y, FHL1-C132F and FHL1-C224W) are not capable of suppressing the FHL1-opathic phenotype in fhl1a–depleted zebrafish embryos. These embryos exhibit severe myopathy with severe functional impairment and morphological disruption, compromised heart function (see Fig. 4E) and pericardial edema (PE). (D) Quantification of the FHL1-opathic phenotype of the injected embryos. The majority of fhl1a-depleted zebrafish embryos expressing mutated FHL1 exhibit FHL1-opathic phenotype. Only human wild-type FHL1 protein is capable of suppressing the FHL1-opathic phenotype. Rate of FHL1-opathic phenotype is 78% ± 1% (n = 130 embryos in total, three independent experiments, p = 0.4457) in MO-fhl1a + FHL1-H123Y, 76% ± 2% (n = 130 embryos in total, three independent experiments, p = 0.6967) in MO-fhl1a + FHL1-C132F and 67% ± 1% (n = 130 embryos in total, three independent experiments, p = 0.4993) in MO-fhl1a + FHL1-C224W, compared to 73% ± 4% (n = 130 embryos in total, three independent experiments) of FHL1-opathic embryos in the fhl1a-morphants and only 34% ± 8% (n = 230 embryos in total, three independent experiments, p = 0.0232) in MO-fhl1a + wild-type FHL1. (E) Cardiac function could not be rescued and heart rate was not normalized in fhl1a-morphants, when mutated human FHL1 is expressed in these morphants. In contrast, heart rate of fhl1a-morphants expressing human wild-type FHL1 normalized. Heart rate remained decreased to 74 ± 8 bpm (n = 20 embryos in total, three independent experiments, p = 0.5918) in MO-fhl1a + FHL1-H123Y, 84 ± 14 bpm (n = 20 embryos in total, three independent experiments, 0.3142) in MO-fhl1a + FHL1-C132F and 70 ± 13 bpm (n = 20 embryos in total, three independent experiments, p = 0.6933) in MO-fhl1a + FHL1-C224W, compared to 89 ± 8 bpm (n = 20 embryos in total, three independent experiments) in fhl1a-morphants and 151 ± 1 bpm (n = 20 embryos in total, three independent experiments) in wild-type control embryos. In contrast, the heart rate of fhl1a-morphants could be normalized to 130 ± 3 bpm (n = 20 embryos in total, three independent experiments; p < 0.0001) by overexpressing human wild-type FHL1.