Fig. 8
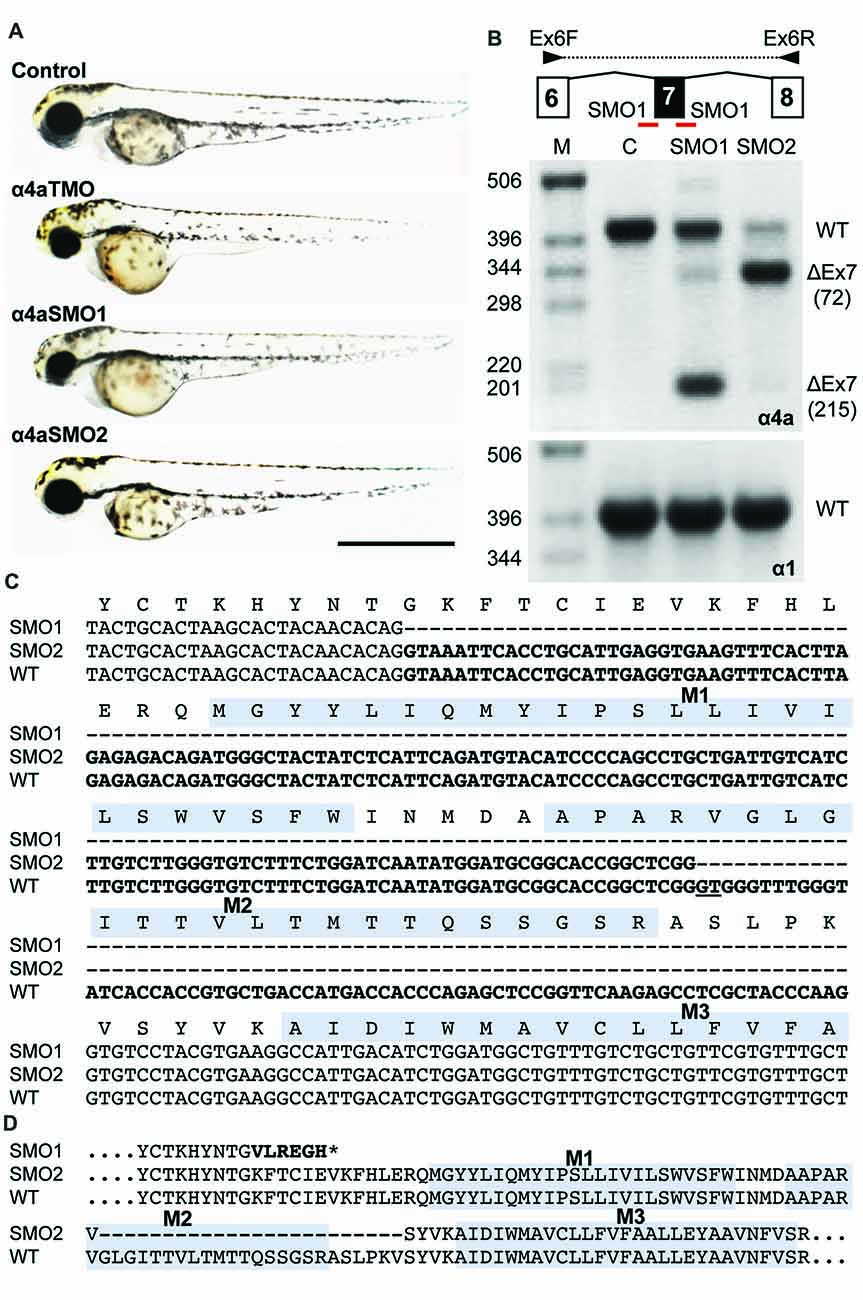
Morpholino oligonucleotides (SMO1 and SMO2) block splicing of glra4a. (A) Gross morphology of wild-type embryos and those injected with GlyR α4a translation-blocking (TMO) or splice site (SMO1 and SMO2) morpholinos. (B) Schematic of RT-PCR analysis of glra4a morpholinos confirmed the deletion of exon 7 for α4a-SMO1 (lane 3) and the deletion of part of exon 7 for α4a-SMO2 (lane 4). Intact GlyR α4a exon 7–9 PCR products are seen in the control (393 bp, lane 2) and smaller amounts are also observed in lanes 3 and 4. A fragment of around 178 bp is seen in SMO1, which is made up of exon 6 and 8 only. A fragment of 321 bp in SMO2 contains exons 6 and 8 and part of exon 7, which excludes the region that codes for TM2. GlyR α1 exon 7 is present in control and both SMO fish (lower panel). (C,D) DNA sequencing of GlyR α4a cDNAs from SMO1 and SMO2 treated fish reveals that SMO1 results in a 215 bp deletion, resulting in a frameshift and premature stop codon before M1, while SMO2 results in a 72 bp deletion and loss of 24 amino acids, including the majority of the pore-forming M2 domain.